The synovial and blood monocyte DNA methylomes mirror prognosis, evolution, and treatment in early arthritis
- PMID: 35324478
- PMCID: PMC9090240
- DOI: 10.1172/jci.insight.158783
The synovial and blood monocyte DNA methylomes mirror prognosis, evolution, and treatment in early arthritis
Abstract
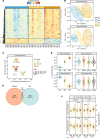
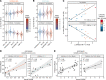
Identifying predictive biomarkers at early stages of inflammatory arthritis is crucial for starting appropriate therapies to avoid poor outcomes. Monocytes (MOs) and macrophages, largely associated with arthritis, are contributors and sensors of inflammation through epigenetic modifications. In this study, we investigated associations between clinical features and DNA methylation in blood and synovial fluid (SF) MOs in a prospective cohort of patients with early inflammatory arthritis. DNA methylation profiles of undifferentiated arthritis (UA) blood MOs exhibited marked alterations in comparison with those from healthy donors. We identified additional differences both in blood and SF MOs after comparing patients with UA grouped by their future outcomes, i.e., good versus poor. Patient profiles in subsequent visits revealed a reversion toward a healthy level in both groups, those requiring disease-modifying antirheumatic drugs and those who remitted spontaneously. Changes in disease activity between visits also affected DNA methylation, which was partially concomitant in the SF of UA and in blood MOs of patients with rheumatoid arthritis. Epigenetic similarities between arthritis types allow a common prediction of disease activity. Our results constitute a resource of DNA methylation-based biomarkers of poor prognosis, disease activity, and treatment efficacy for the personalized clinical management of early inflammatory arthritis.
Keywords: Arthritis; Autoimmunity; Epigenetics; Inflammation.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

