Genetic Analysis of Influenza A/H1N1pdm Strains Isolated in Bangladesh in Early 2020
- PMID: 35324585
- PMCID: PMC8949303
- DOI: 10.3390/tropicalmed7030038
Genetic Analysis of Influenza A/H1N1pdm Strains Isolated in Bangladesh in Early 2020
Abstract
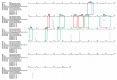
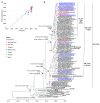
Influenza is one of the most common respiratory virus infections. We analyzed hemagglutinin (HA) and neuraminidase (NA) gene segments of viruses isolated from influenza patients who visited Evercare Hospital Dhaka, Bangladesh, in early 2020 immediately before the coronavirus disease 2019 (COVID-19) pandemic. All of them were influenza virus type A (IAV) H1N1pdm. Sequence analysis of the HA segments of the virus strains isolated from the clinical specimens and the subsequent phylogenic analyses of the obtained sequences revealed that all of the H1N1pdm recent subclades 6B.1A5A + 187V/A, 6B.1A5A + 156K, and 6B.1A5A + 156K with K209M were already present in Bangladesh in January 2020. Molecular clock analysis results suggested that the subclade 6B.1A5A + 156K emerged in Denmark, Australia, or the United States in July 2019, while subclades 6B.1A5A + 187V/A and 6B.1A5A + 156K with K209M emerged in East Asia in April and September 2019, respectively. On the other hand, sequence analysis of NA segments showed that the viruses lacked the H275Y mutation that confers oseltamivir resistance. Since the number of influenza cases in Bangladesh is usually small between November and January, these results indicated that the IAV H1N1pdm had spread extremely rapidly without acquiring oseltamivir resistance during a time of active international flow of people before the COVID-19 pandemic.
Keywords: Bangladesh; COVID-19; H1N1pdm09; HA; NA; evolution; influenza virus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Cluster of Oseltamivir-Resistant and Hemagglutinin Antigenically Drifted Influenza A(H1N1)pdm09 Viruses, Texas, USA, January 2020.Emerg Infect Dis. 2021 Jul;27(7):1953-1957. doi: 10.3201/eid2707.204593. Emerg Infect Dis. 2021. PMID: 34152954 Free PMC article.
-
Spatiotemporal Distribution and Evolution of the A/H1N1 2009 Pandemic Influenza Virus in Pigs in France from 2009 to 2017: Identification of a Potential Swine-Specific Lineage.J Virol. 2018 Nov 27;92(24):e00988-18. doi: 10.1128/JVI.00988-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258006 Free PMC article.
-
Phylogenetic analysis of the neuraminidase gene of pandemic H1N1 influenza A virus circulating in the South American region.Virus Res. 2015 Feb 2;197:1-7. doi: 10.1016/j.virusres.2014.11.008. Epub 2014 Dec 3. Virus Res. 2015. PMID: 25479596
-
Rapid discrimination of oseltamivir-resistant 275Y and -susceptible 275H substitutions in the neuraminidase gene of pandemic influenza A/H1N1 2009 virus by duplex one-step RT-PCR assay.J Med Virol. 2011 Jul;83(7):1121-7. doi: 10.1002/jmv.22101. J Med Virol. 2011. PMID: 21567417 Free PMC article.
-
Amino acids transitioning of 2009 H1N1pdm in Taiwan from 2009 to 2011.PLoS One. 2012;7(9):e45946. doi: 10.1371/journal.pone.0045946. Epub 2012 Sep 24. PLoS One. 2012. PMID: 23029335 Free PMC article.
Cited by
-
Mutation in Hemagglutinin Antigenic Sites in Influenza A pH1N1 Viruses from 2015-2019 in the United States Mountain West, Europe, and the Northern Hemisphere.Genes (Basel). 2022 May 19;13(5):909. doi: 10.3390/genes13050909. Genes (Basel). 2022. PMID: 35627294 Free PMC article.
-
Tropical Infectious Diseases of Global Significance: Insights and Perspectives.Trop Med Infect Dis. 2023 Sep 29;8(10):462. doi: 10.3390/tropicalmed8100462. Trop Med Infect Dis. 2023. PMID: 37888590 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

