Microbiome in Chronic Kidney Disease (CKD): An Omics Perspective
- PMID: 35324673
- PMCID: PMC8951538
- DOI: 10.3390/toxins14030176
Microbiome in Chronic Kidney Disease (CKD): An Omics Perspective
Abstract
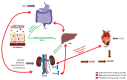
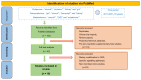
Chronic kidney disease (CKD) is predominant in 10% of the world's adult population, and is increasingly considered a silent epidemic. Gut microbiota plays an essential role in maintaining host energy homeostasis and gut epithelial integrity. Alterations in gut microbiota composition, functions and, specifically, production of metabolites causing uremic toxicity are often associated with CKD onset and progression. Here, we present the latest omics (transcriptomics, proteomics and metabolomics) studies that explore the connection between CKD and gut microbiome. A review of the available literature using PubMed was performed using the keywords "microb*", "kidney", "proteom", "metabolom" and "transcript" for the last 10 years, yielding a total of 155 publications. Following selection of the relevant studies (focusing on microbiome in CKD), a predominance of metabolomics (n = 12) over transcriptomics (n = 1) and proteomics (n = 6) analyses was observed. A consensus arises supporting the idea that the uremic toxins produced in the gut cause oxidative stress, inflammation and fibrosis in the kidney leading to CKD. Collectively, findings include an observed enrichment of Eggerthella lenta, Enterobacteriaceae and Clostridium spp., and a depletion in Bacteroides eggerthii, Roseburia faecis and Prevotella spp. occurring in CKD models. Bacterial species involved in butyrate production, indole synthesis and mucin degradation were also related to CKD. Consequently, strong links between CKD and gut microbial dysbiosis suggest potential therapeutic strategies to prevent CKD progression and portray the gut as a promising therapeutic target.
Keywords: CKD; gut microbiota; omics; therapeutic targets; uremic toxins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

