Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans
- PMID: 35324723
- PMCID: PMC8949452
- DOI: 10.3390/toxins14030226
Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus distans
Abstract
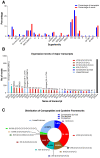
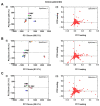
The defensive use of cone snail venom is hypothesised to have first arisen in ancestral worm-hunting snails and later repurposed in a compartmentalised venom duct to facilitate the dietary shift to molluscivory and piscivory. Consistent with its placement in a basal lineage, we demonstrate that the C. distans venom gland lacked distinct compartmentalisation. Transcriptomics revealed C. distans expressed a wide range of structural classes, with inhibitory cysteine knot (ICK)-containing peptides dominating. To better understand the evolution of the venom gland compartmentalisation, we compared C. distans to C. planorbis, the earliest diverging species from which a defence-evoked venom has been obtained, and fish-hunting C. geographus from the Gastridium subgenus that injects distinct defensive and predatory venoms. These comparisons support the hypothesis that venom gland compartmentalisation arose in worm-hunting species and enabled repurposing of venom peptides to facilitate the dietary shift from vermivory to molluscivory and piscivory in more recently diverged cone snail lineages.
Keywords: conotoxins; defensive venom; evolution; proteomics; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Predatory and Defensive Strategies in Cone Snails.Toxins (Basel). 2024 Feb 7;16(2):94. doi: 10.3390/toxins16020094. Toxins (Basel). 2024. PMID: 38393171 Free PMC article. Review.
-
The role of defensive ecological interactions in the evolution of conotoxins.Mol Ecol. 2016 Jan;25(2):598-615. doi: 10.1111/mec.13504. Epub 2016 Jan 20. Mol Ecol. 2016. PMID: 26614983
-
Venom duct origins of prey capture and defensive conotoxins in piscivorous Conus striatus.Sci Rep. 2021 Jun 24;11(1):13282. doi: 10.1038/s41598-021-91919-4. Sci Rep. 2021. PMID: 34168165 Free PMC article.
-
Diversity and Novelty of Venom Peptides in Vermivorous Cone Snails, Subgenus Rhizoconus (Gastropoda: Mollusca).Mar Drugs. 2025 Jun 26;23(7):266. doi: 10.3390/md23070266. Mar Drugs. 2025. PMID: 40710491 Free PMC article.
-
Venomics-Accelerated Cone Snail Venom Peptide Discovery.Int J Mol Sci. 2018 Mar 9;19(3):788. doi: 10.3390/ijms19030788. Int J Mol Sci. 2018. PMID: 29522462 Free PMC article. Review.
Cited by
-
Potential Ancestral Conoidean Toxins in the Venom Cocktail of the Carnivorous Snail Raphitoma purpurea (Montagu, 1803) (Neogastropoda: Raphitomidae).Toxins (Basel). 2024 Aug 9;16(8):348. doi: 10.3390/toxins16080348. Toxins (Basel). 2024. PMID: 39195758 Free PMC article.
-
Predatory and Defensive Strategies in Cone Snails.Toxins (Basel). 2024 Feb 7;16(2):94. doi: 10.3390/toxins16020094. Toxins (Basel). 2024. PMID: 38393171 Free PMC article. Review.
References
-
- Undheim E.A., Hamilton B.R., Kurniawan N.D., Bowlay G., Cribb B.W., Merritt D.J., Fry B.G., King G.F., Venter D.J. Production and packaging of a biological arsenal: Evolution of centipede venoms under morphological constraint. Proc. Natl. Acad. Sci. USA. 2015;112:4026–4031. doi: 10.1073/pnas.1424068112. - DOI - PMC - PubMed
-
- Fry B.G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ramjan S.F.R., Vidal N., Poelmann R.E., Norman J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia) Mol. Cell. Proteom. 2008;7:215–246. doi: 10.1074/mcp.M700094-MCP200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

