Isotope-assisted metabolic flux analysis as an equality-constrained nonlinear program for improved scalability and robustness
- PMID: 35324890
- PMCID: PMC8947808
- DOI: 10.1371/journal.pcbi.1009831
Isotope-assisted metabolic flux analysis as an equality-constrained nonlinear program for improved scalability and robustness
Abstract
Stable isotope-assisted metabolic flux analysis (MFA) is a powerful method to estimate carbon flow and partitioning in metabolic networks. At its core, MFA is a parameter estimation problem wherein the fluxes and metabolite pool sizes are model parameters that are estimated, via optimization, to account for measurements of steady-state or isotopically-nonstationary isotope labeling patterns. As MFA problems advance in scale, they require efficient computational methods for fast and robust convergence. The structure of the MFA problem enables it to be cast as an equality-constrained nonlinear program (NLP), where the equality constraints are constructed from the MFA model equations, and the objective function is defined as the sum of squared residuals (SSR) between the model predictions and a set of labeling measurements. This NLP can be solved by using an algebraic modeling language (AML) that offers state-of-the-art optimization solvers for robust parameter estimation and superior scalability to large networks. When implemented in this manner, the optimization is performed with no distinction between state variables and model parameters. During each iteration of such an optimization, the system state is updated instead of being calculated explicitly from scratch, and this occurs concurrently with improvement in the model parameter estimates. This optimization approach starkly contrasts with traditional "shooting" methods where the state variables and model parameters are kept distinct and the system state is computed afresh during each iteration of a stepwise optimization. Our NLP formulation uses the MFA modeling framework of Wiechert et al. [1], which is amenable to incorporation of the model equations into an NLP. The NLP constraints consist of balances on either elementary metabolite units (EMUs) or cumomers. In this formulation, both the steady-state and isotopically-nonstationary MFA (inst-MFA) problems may be solved as an NLP. For the inst-MFA case, the ordinary differential equation (ODE) system describing the labeling dynamics is transcribed into a system of algebraic constraints for the NLP using collocation. This large-scale NLP may be solved efficiently using an NLP solver implemented on an AML. In our implementation, we used the reduced gradient solver CONOPT, implemented in the General Algebraic Modeling System (GAMS). The NLP framework is particularly advantageous for inst-MFA, scaling well to large networks with many free parameters, and having more robust convergence properties compared to the shooting methods that compute the system state and sensitivities at each iteration. Additionally, this NLP approach supports the use of tandem-MS data for both steady-state and inst-MFA when the cumomer framework is used. We assembled a software, eiFlux, written in Python and GAMS that uses the NLP approach and supports both steady-state and inst-MFA. We demonstrate the effectiveness of the NLP formulation on several examples, including a genome-scale inst-MFA model, to highlight the scalability and robustness of this approach. In addition to typical inst-MFA applications, we expect that this framework and our associated software, eiFlux, will be particularly useful for applying inst-MFA to complex MFA models, such as those developed for eukaryotes (e.g. algae) and co-cultures with multiple cell types.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


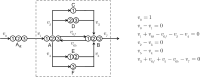
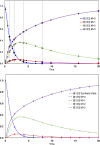
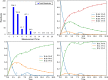
Similar articles
-
Isotopically nonstationary metabolic flux analysis (INST-MFA): putting theory into practice.Curr Opin Biotechnol. 2018 Dec;54:80-87. doi: 10.1016/j.copbio.2018.02.013. Epub 2018 Mar 6. Curr Opin Biotechnol. 2018. PMID: 29522915 Review.
-
Pool size measurements improve precision of flux estimates but increase sensitivity to unmodeled reactions outside the core network in isotopically nonstationary metabolic flux analysis (INST-MFA).Biotechnol J. 2022 Mar;17(3):e2000427. doi: 10.1002/biot.202000427. Epub 2022 Feb 11. Biotechnol J. 2022. PMID: 35085426
-
A simulation-free constrained regression approach for flux estimation in isotopically nonstationary metabolic flux analysis with applications in microalgae.Front Plant Sci. 2023 Nov 23;14:1140829. doi: 10.3389/fpls.2023.1140829. eCollection 2023. Front Plant Sci. 2023. PMID: 38078077 Free PMC article.
-
Isotopically nonstationary MFA (INST-MFA) of autotrophic metabolism.Methods Mol Biol. 2014;1090:181-210. doi: 10.1007/978-1-62703-688-7_12. Methods Mol Biol. 2014. PMID: 24222417
-
Isotopically non-stationary metabolic flux analysis: complex yet highly informative.Curr Opin Biotechnol. 2013 Dec;24(6):979-86. doi: 10.1016/j.copbio.2013.03.024. Epub 2013 Apr 24. Curr Opin Biotechnol. 2013. PMID: 23623747 Review.
Cited by
-
DIMet: an open-source tool for differential analysis of targeted isotope-labeled metabolomics data.Bioinformatics. 2024 May 2;40(5):btae282. doi: 10.1093/bioinformatics/btae282. Bioinformatics. 2024. PMID: 38656970 Free PMC article.
-
Systematic comparison of local approaches for isotopically nonstationary metabolic flux analysis.Front Plant Sci. 2023 Jun 6;14:1178239. doi: 10.3389/fpls.2023.1178239. eCollection 2023. Front Plant Sci. 2023. PMID: 37346134 Free PMC article.
-
Metabolic flux analysis in adipose tissue reprogramming.Immunometabolism (Cobham). 2024 Mar 6;6(1):e00039. doi: 10.1097/IN9.0000000000000039. eCollection 2024 Jan. Immunometabolism (Cobham). 2024. PMID: 38455681 Free PMC article. Review.
-
Metabolomics-driven approaches for identifying therapeutic targets in drug discovery.MedComm (2020). 2024 Nov 11;5(11):e792. doi: 10.1002/mco2.792. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39534557 Free PMC article. Review.
-
Interpreting metabolic complexity via isotope-assisted metabolic flux analysis.Trends Biochem Sci. 2023 Jun;48(6):553-567. doi: 10.1016/j.tibs.2023.02.001. Epub 2023 Mar 1. Trends Biochem Sci. 2023. PMID: 36863894 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

