Genome wide association study of Escherichia coli bloodstream infection isolates identifies genetic determinants for the portal of entry but not fatal outcome
- PMID: 35324915
- PMCID: PMC8946752
- DOI: 10.1371/journal.pgen.1010112
Genome wide association study of Escherichia coli bloodstream infection isolates identifies genetic determinants for the portal of entry but not fatal outcome
Abstract
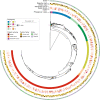
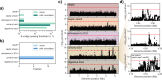
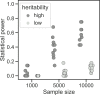
Escherichia coli is an important cause of bloodstream infections (BSI), which is of concern given its high mortality and increasing worldwide prevalence. Finding bacterial genetic variants that might contribute to patient death is of interest to better understand infection progression and implement diagnostic methods that specifically look for those factors. E. coli samples isolated from patients with BSI are an ideal dataset to systematically search for those variants, as long as the influence of host factors such as comorbidities are taken into account. Here we performed a genome-wide association study (GWAS) using data from 912 patients with E. coli BSI from hospitals in Paris, France. We looked for associations between bacterial genetic variants and three patient outcomes (death at 28 days, septic shock and admission to intensive care unit), as well as two portals of entry (urinary and digestive tract), using various clinical variables from each patient to account for host factors. We did not find any association between genetic variants and patient outcomes, potentially confirming the strong influence of host factors in influencing the course of BSI; we however found a strong association between the papGII operon and entrance of E. coli through the urinary tract, which demonstrates the power of bacterial GWAS when applied to actual clinical data. Despite the lack of associations between E. coli genetic variants and patient outcomes, we estimate that increasing the sample size by one order of magnitude could lead to the discovery of some putative causal variants. Given the wide adoption of bacterial genome sequencing of clinical isolates, such sample sizes may be soon available.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Mortality in Escherichia coli bloodstream infections: antibiotic resistance still does not make it.J Antimicrob Chemother. 2020 Aug 1;75(8):2334-2343. doi: 10.1093/jac/dkaa161. J Antimicrob Chemother. 2020. PMID: 32417924
-
A 21-Year Survey of Escherichia coli from Bloodstream Infections (BSI) in a Tertiary Hospital Reveals How Community-Hospital Dynamics of B2 Phylogroup Clones Influence Local BSI Rates.mSphere. 2021 Dec 22;6(6):e0086821. doi: 10.1128/msphere.00868-21. Epub 2021 Dec 22. mSphere. 2021. PMID: 34935444 Free PMC article.
-
Bacterial genome-wide association study substantiates papGII of Escherichia coli as a major risk factor for urosepsis.Genome Med. 2023 Oct 30;15(1):89. doi: 10.1186/s13073-023-01243-x. Genome Med. 2023. PMID: 37904175 Free PMC article.
-
Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis.Clin Microbiol Infect. 2007 Sep;13(9):854-62. doi: 10.1111/j.1469-0691.2007.01775.x. Epub 2007 Jul 6. Clin Microbiol Infect. 2007. PMID: 17617183
-
Epidemiology and impact of bloodstream infections among kidney transplant recipients: A retrospective single-center experience.Transpl Infect Dis. 2018 Feb;20(1). doi: 10.1111/tid.12815. Epub 2018 Jan 19. Transpl Infect Dis. 2018. PMID: 29151282 Review.
Cited by
-
Word-based GWAS harnesses the rich potential of genomic data for E. coli quinolone resistance.Front Microbiol. 2023 Dec 13;14:1276332. doi: 10.3389/fmicb.2023.1276332. eCollection 2023. Front Microbiol. 2023. PMID: 38152371 Free PMC article.
-
Evolutionary genomic analyses of canine E. coli infections identify a relic capsular locus associated with resistance to multiple classes of antimicrobials.Appl Environ Microbiol. 2024 Aug 21;90(8):e0035424. doi: 10.1128/aem.00354-24. Epub 2024 Jul 16. Appl Environ Microbiol. 2024. PMID: 39012166 Free PMC article.
-
Predicting the primary infection source of Escherichia coli bacteremia using virulence-associated genes.Eur J Clin Microbiol Infect Dis. 2024 Apr;43(4):641-648. doi: 10.1007/s10096-024-04754-6. Epub 2024 Jan 25. Eur J Clin Microbiol Infect Dis. 2024. PMID: 38273191 Free PMC article.
-
Staphylococcus aureus host interactions and adaptation.Nat Rev Microbiol. 2023 Jun;21(6):380-395. doi: 10.1038/s41579-023-00852-y. Epub 2023 Jan 27. Nat Rev Microbiol. 2023. PMID: 36707725 Free PMC article. Review.
-
All Staphylococcus aureus bacteraemia-inducing strains can cause infective endocarditis: Results of GWAS and experimental animal studies.J Infect. 2023 Feb;86(2):123-133. doi: 10.1016/j.jinf.2022.12.028. Epub 2023 Jan 2. J Infect. 2023. PMID: 36603774 Free PMC article.
References
-
- Yoon E-J, Choi MH, Park YS, Lee HS, Kim D, Lee H, et al.. Impact of host-pathogen-treatment tripartite components on early mortality of patients with Escherichia coli bloodstream infection: Prospective observational study. EBioMedicine. 2018;35: 76–86. doi: 10.1016/j.ebiom.2018.08.029 - DOI - PMC - PubMed
-
- MacKinnon MC, McEwen SA, Pearl DL, Lyytikäinen O, Jacobsson G, Collignon P, et al.. Increasing incidence and antimicrobial resistance in Escherichia coli bloodstream infections: a multinational population-based cohort study. Antimicrob Resist Infect Control. 2021;10: 131. doi: 10.1186/s13756-021-00999-4 - DOI - PMC - PubMed
-
- Desjardins P, Picard B, Kaltenböck B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41: 440–448. doi: 10.1007/BF00160315 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

