Genetic code degeneracy is established by the decoding center of the ribosome
- PMID: 35325219
- PMCID: PMC9023292
- DOI: 10.1093/nar/gkac171
Genetic code degeneracy is established by the decoding center of the ribosome
Abstract
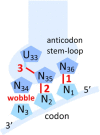
The degeneracy of the genetic code confers a wide array of properties to coding sequences. Yet, its origin is still unclear. A structural analysis has shown that the stability of the Watson-Crick base pair at the second position of the anticodon-codon interaction is a critical parameter controlling the extent of non-specific pairings accepted at the third position by the ribosome, a flexibility at the root of degeneracy. Based on recent cryo-EM analyses, the present work shows that residue A1493 of the decoding center provides a significant contribution to the stability of this base pair, revealing that the ribosome is directly involved in the establishment of degeneracy. Building on existing evolutionary models, we show the evidence that the early appearance of A1493 and A1492 established the basis of degeneracy when an elementary kinetic scheme of translation was prevailing. Logical considerations on the expansion of this kinetic scheme indicate that the acquisition of the peptidyl transferase center was the next major evolutionary step, while the induced-fit mechanism, that enables a sharp selection of the tRNAs, necessarily arose later when G530 was acquired by the decoding center.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon.RNA. 2008 Jul;14(7):1264-9. doi: 10.1261/rna.1029808. Epub 2008 May 21. RNA. 2008. PMID: 18495942 Free PMC article.
-
Testing constraints on rRNA bases that make nonsequence-specific contacts with the codon-anticodon complex in the ribosomal A site.RNA. 2007 Aug;13(8):1279-86. doi: 10.1261/rna.552007. Epub 2007 Jun 25. RNA. 2007. PMID: 17592040 Free PMC article.
-
The ribosome's response to codon-anticodon mismatches.Biochimie. 2006 Aug;88(8):1001-11. doi: 10.1016/j.biochi.2006.04.013. Epub 2006 May 12. Biochimie. 2006. PMID: 16716484 Review.
-
Steric complementarity in the decoding center is important for tRNA selection by the ribosome.J Mol Biol. 2013 Oct 23;425(20):3778-89. doi: 10.1016/j.jmb.2013.02.038. Epub 2013 Mar 27. J Mol Biol. 2013. PMID: 23542008 Free PMC article.
-
Two distinct conformations of the conserved RNA-rich decoding center of the small ribosomal subunit are recognized by tRNAs and release factors.Cold Spring Harb Symp Quant Biol. 2006;71:545-9. doi: 10.1101/sqb.2006.71.036. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381338 Review.
Cited by
-
Initial Amino Acid:Codon Assignments and Strength of Codon:Anticodon Binding.J Am Chem Soc. 2024 May 8;146(18):12857-12863. doi: 10.1021/jacs.4c03644. Epub 2024 Apr 27. J Am Chem Soc. 2024. PMID: 38676654 Free PMC article.
-
Comprehensive translational profiling and STE AI uncover rapid control of protein biosynthesis during cell stress.Nucleic Acids Res. 2024 Jul 22;52(13):7925-7946. doi: 10.1093/nar/gkae365. Nucleic Acids Res. 2024. PMID: 38721779 Free PMC article.
-
Genomic AT Bias Coupled with Amino Acid Metabolism Modulates Codon Usage.J Mol Evol. 2025 Jun;93(3):379-394. doi: 10.1007/s00239-025-10251-x. Epub 2025 May 20. J Mol Evol. 2025. PMID: 40392286
-
Genome-wide impact of codon usage bias on translation optimization in Drosophila melanogaster.Nat Commun. 2024 Sep 27;15(1):8329. doi: 10.1038/s41467-024-52660-4. Nat Commun. 2024. PMID: 39333102 Free PMC article.
References
-
- Nirenberg M. Historical review: deciphering the genetic code–a personal account. Trends Biochem. Sci. 2004; 29:46–54. - PubMed
-
- Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. - PubMed
-
- Ogle J.M., Brodersen D.E., Clemons W.M.J., Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. - PubMed
-
- Ogle J.M., Murphy F.V, Tarry M.J., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. - PubMed
-
- Ogle J.M., Ramakrishnan V.. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005; 74:129–177. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

