Correlation Imputation for Single-Cell RNA-seq
- PMID: 35325552
- PMCID: PMC9125575
- DOI: 10.1089/cmb.2021.0403
Correlation Imputation for Single-Cell RNA-seq
Abstract
Recent advances in single-cell RNA sequencing (scRNA-seq) technologies have yielded a powerful tool to measure gene expression of individual cells. One major challenge of the scRNA-seq data is that it usually contains a large amount of zero expression values, which often impairs the effectiveness of downstream analyses. Numerous data imputation methods have been proposed to deal with these "dropout" events, but this is a difficult task for such high-dimensional and sparse data. Furthermore, there have been debates on the nature of the sparsity, about whether the zeros are due to technological limitations or represent actual biology. To address these challenges, we propose Single-cell RNA-seq Correlation completion by ENsemble learning and Auxiliary information (SCENA), a novel approach that imputes the correlation matrix of the data of interest instead of the data itself. SCENA obtains a gene-by-gene correlation estimate by ensembling various individual estimates, some of which are based on known auxiliary information about gene expression networks. Our approach is a reliable method that makes no assumptions on the nature of sparsity in scRNA-seq data or the data distribution. By extensive simulation studies and real data applications, we demonstrate that SCENA is not only superior in gene correlation estimation, but also improves the accuracy and reliability of downstream analyses, including cell clustering, dimension reduction, and graphical model estimation to learn the gene expression network.
Keywords: auxiliary information; clustering; correlation completion; dimension reduction; ensemble learning; graphical modeling; imputation; single-cell RNA-sequencing.
Conflict of interest statement
The authors declare they have no conflicting financial interests.
Figures
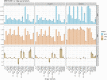


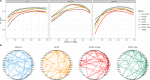



References
-
- Chen, C., Wu, C., Wu, L., et al. . 2018. scRMD: Imputation for single cell RNA-seq data via robust matrix decomposition. bioRxiv 459404. Bioinformatics. 36, 3156–3161. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

