The E3 Ubiquitin Ligase Gene Sl1 Is Critical for Cadmium Tolerance in Solanum lycopersicum L
- PMID: 35326106
- PMCID: PMC8944816
- DOI: 10.3390/antiox11030456
The E3 Ubiquitin Ligase Gene Sl1 Is Critical for Cadmium Tolerance in Solanum lycopersicum L
Abstract
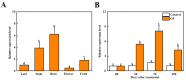
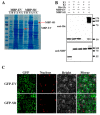
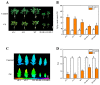
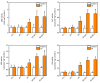
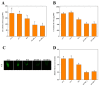
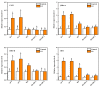
Heavy metal cadmium (Cd) at high concentrations severely disturbs plant growth and development. The E3 ubiquitin ligase involved in protein degradation is critical for plant tolerance to abiotic stress, but the role of E3 ubiquitin ligases in Cd tolerance is largely unknown in tomato. Here, we characterized an E3 ubiquitin ligase gene Sl1, which was highly expressed in roots under Cd stress in our previous study. The subcellular localization of Sl1 revealed that it was located in plasma membranes. In vitro ubiquitination assays confirmed that Sl1 had E3 ubiquitin ligase activity. Knockout of the Sl1 gene by CRISPR/Cas9 genome editing technology reduced while its overexpression increased Cd tolerance as reflected by the changes in the actual quantum efficiency of PSII photochemistry (ΦPSII) and hydrogen peroxide (H2O2) accumulation. Cd-induced increased activities of antioxidant enzymes including superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and glutathione reductase (GR) were compromised in sl1 mutants but were enhanced in Sl1 overexpressing lines. Furthermore, the content of Cd in both shoots and roots increased in sl1 mutants while reduced in Sl1 overexpressing plants. Gene expression assays revealed that Sl1 regulated the transcript levels of heavy metal transport-related genes to inhibit Cd accumulation. These findings demonstrate that Sl1 plays a critical role in regulating Cd tolerance by relieving oxidative stress and resisting heavy metal transportation in tomato. The study provides a new understanding of the mechanism of plant tolerance to heavy metal stress.
Keywords: antioxidant enzymes; heavy metal stress; protein degradation; tomato; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum.J Biotechnol. 2020 Dec 20;324:239-247. doi: 10.1016/j.jbiotec.2020.11.008. Epub 2020 Nov 10. J Biotechnol. 2020. PMID: 33186659
-
Overexpression of tomato RING E3 ubiquitin ligase gene SlRING1 confers cadmium tolerance by attenuating cadmium accumulation and oxidative stress.Physiol Plant. 2021 Sep;173(1):449-459. doi: 10.1111/ppl.13294. Epub 2020 Dec 16. Physiol Plant. 2021. PMID: 33616963
-
Insights into citric acid-induced cadmium tolerance and phytoremediation in Brassica juncea L.: Coordinated functions of metal chelation, antioxidant defense and glyoxalase systems.Ecotoxicol Environ Saf. 2018 Jan;147:990-1001. doi: 10.1016/j.ecoenv.2017.09.045. Epub 2017 Oct 7. Ecotoxicol Environ Saf. 2018. PMID: 29976011
-
Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization.J Exp Bot. 2002 May;53(372):1351-65. J Exp Bot. 2002. PMID: 11997381 Review.
-
[Mechanisms of heavy metal cadmium tolerance in plants].Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2006 Feb;32(1):1-8. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2006. PMID: 16477124 Review. Chinese.
Cited by
-
The Potential of CRISPR/Cas Technology to Enhance Crop Performance on Adverse Soil Conditions.Plants (Basel). 2023 May 5;12(9):1892. doi: 10.3390/plants12091892. Plants (Basel). 2023. PMID: 37176948 Free PMC article. Review.
-
The role of CBL-CIPK signaling in plant responses to biotic and abiotic stresses.Plant Mol Biol. 2024 May 7;114(3):53. doi: 10.1007/s11103-024-01417-0. Plant Mol Biol. 2024. PMID: 38714550 Review.
-
Molecular Mechanisms of Cadmium Stress Resistance in Vegetable Crops.Int J Mol Sci. 2025 Jun 17;26(12):5812. doi: 10.3390/ijms26125812. Int J Mol Sci. 2025. PMID: 40565274 Free PMC article. Review.
-
Defense guard: strategies of plants in the fight against Cadmium stress.Adv Biotechnol (Singap). 2024 Dec 2;2(4):44. doi: 10.1007/s44307-024-00052-6. Adv Biotechnol (Singap). 2024. PMID: 39883385 Free PMC article. Review.
-
Transcriptome Analysis Reveals Changes in Whole Gene Expression, Biological Process, and Molecular Functions Induced by Nickel in Jack Pine (Pinus banksiana).Plants (Basel). 2023 Aug 7;12(15):2889. doi: 10.3390/plants12152889. Plants (Basel). 2023. PMID: 37571042 Free PMC article.
References
-
- Zhang G., Fukami M., Sekimoto H. Genotypic differences in effects of cadmium on growth and nutrient compositions in wheat. J. Plant Nutr. 2000;23:1337–1350. doi: 10.1080/01904160009382104. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

