Analyzing the Androgen Receptor Interactome in Prostate Cancer: Implications for Therapeutic Intervention
- PMID: 35326387
- PMCID: PMC8946651
- DOI: 10.3390/cells11060936
Analyzing the Androgen Receptor Interactome in Prostate Cancer: Implications for Therapeutic Intervention
Abstract
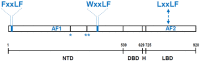
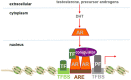
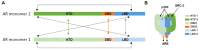
The androgen receptor (AR) is a member of the ligand-activated nuclear receptor family of transcription factors. AR's transactivation activity is turned on by the binding of androgens, the male sex steroid hormones. AR is critical for the development and maintenance of the male phenotype but has been recognized to also play an important role in human diseases. Most notably, AR is a major driver of prostate cancer (CaP) progression, which remains the second leading cause of cancer deaths in American men. Androgen deprivation therapies (ADTs) that interfere with interactions between AR and its activating androgen ligands have been the mainstay for treatment of metastatic CaP. Although ADTs are effective and induce remissions, eventually they fail, while the growth of the majority of ADT-resistant CaPs remains under AR's control. Alternative approaches to inhibit AR activity and bypass resistance to ADT are being sought, such as preventing the interaction between AR and its cofactors and coregulators that is needed to execute AR-dependent transcription. For such strategies to be efficient, the 3D conformation of AR complexes needs to be well-understood and AR-regulator interaction sites resolved. Here, we review current insights into these 3D structures and the protein interaction sites in AR transcriptional complexes. We focus on methods and technological approaches used to identify AR interactors and discuss challenges and limitations that need to be overcome for efficient therapeutic AR complex disruption.
Keywords: androgen deprivation therapy; coactivators; corepressors; hormonal therapy; proteomics; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Androgen action in the prostate gland.Minerva Urol Nefrol. 2012 Mar;64(1):35-49. Minerva Urol Nefrol. 2012. PMID: 22402316 Review.
-
Targeting the androgen receptor signaling pathway in advanced prostate cancer.Am J Health Syst Pharm. 2022 Jul 22;79(15):1224-1235. doi: 10.1093/ajhp/zxac105. Am J Health Syst Pharm. 2022. PMID: 35390118 Review.
-
Functional implications and therapeutic targeting of androgen response elements in prostate cancer.Biochimie. 2023 Nov;214(Pt B):188-198. doi: 10.1016/j.biochi.2023.07.012. Epub 2023 Jul 17. Biochimie. 2023. PMID: 37460038 Review.
-
Alternative approaches to prevent androgen action in prostate cancer: are we there yet?Discov Med. 2014 May;17(95):267-74. Discov Med. 2014. PMID: 24882718
-
AR-dependent phosphorylation and phospho-proteome targets in prostate cancer.Endocr Relat Cancer. 2020 Jun;27(6):R193-R210. doi: 10.1530/ERC-20-0048. Endocr Relat Cancer. 2020. PMID: 32276264 Free PMC article. Review.
Cited by
-
Cellular specificity of androgen receptor, coregulators, and pioneer factors in prostate cancer.Endocr Oncol. 2022 Sep 8;2(1):R112-R131. doi: 10.1530/EO-22-0065. eCollection 2022 Jan. Endocr Oncol. 2022. PMID: 37435460 Free PMC article. Review.
-
Androgen receptor signalling in non-prostatic malignancies: challenges and opportunities.Nat Rev Cancer. 2025 Feb;25(2):93-108. doi: 10.1038/s41568-024-00772-w. Epub 2024 Nov 25. Nat Rev Cancer. 2025. PMID: 39587300 Free PMC article. Review.
-
A Multivalent Peptoid Conjugate Modulates Androgen Receptor Transcriptional Activity to Inhibit Therapy-resistant Prostate Cancer.Mol Cancer Ther. 2023 Oct 2;22(10):1166-1181. doi: 10.1158/1535-7163.MCT-23-0196. Mol Cancer Ther. 2023. PMID: 37486978 Free PMC article.
-
Beyond Prostate Cancer: An Androgen Receptor Splice Variant Expression in Multiple Malignancies, Non-Cancer Pathologies, and Development.Biomedicines. 2023 Aug 7;11(8):2215. doi: 10.3390/biomedicines11082215. Biomedicines. 2023. PMID: 37626712 Free PMC article. Review.
-
Phloretin in Benign Prostate Hyperplasia and Prostate Cancer: A Contemporary Systematic Review.Life (Basel). 2022 Jul 11;12(7):1029. doi: 10.3390/life12071029. Life (Basel). 2022. PMID: 35888117 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

