Batch Effects during Human Bone Marrow Stromal Cell Propagation Prevail Donor Variation and Culture Duration: Impact on Genotype, Phenotype and Function
- PMID: 35326396
- PMCID: PMC8946746
- DOI: 10.3390/cells11060946
Batch Effects during Human Bone Marrow Stromal Cell Propagation Prevail Donor Variation and Culture Duration: Impact on Genotype, Phenotype and Function
Abstract
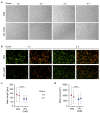
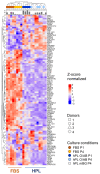
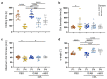
Donor variation is a prominent critical issue limiting the applicability of cell-based therapies. We hypothesized that batch effects during propagation of bone marrow stromal cells (BMSCs) in human platelet lysate (hPL), replacing fetal bovine serum (FBS), can affect phenotypic and functional variability. We therefore investigated the impact of donor variation, hPL- vs. FBS-driven propagation and exhaustive proliferation, on BMSC epigenome, transcriptome, phenotype, coagulation risk and osteochondral regenerative function. Notably, propagation in hPL significantly increased BMSC proliferation, created significantly different gene expression trajectories and distinct surface marker signatures, already after just one passage. We confirmed significantly declining proliferative potential in FBS-expanded BMSC after proliferative challenge. Flow cytometry verified the canonical fibroblastic phenotype in culture-expanded BMSCs. We observed limited effects on DNA methylation, preferentially in FBS-driven cultures, irrespective of culture duration. The clotting risk increased over culture time. Moreover, expansion in xenogenic serum resulted in significant loss of function during 3D cartilage disk formation and significantly increased clotting risk. Superior chondrogenic function under hPL-conditions was maintained over culture. The platelet blood group and isoagglutinins had minor impact on BMSC function. These data demonstrate pronounced batch effects on BMSC transcriptome, phenotype and function due to serum factors, partly outcompeting donor variation after just one culture passage.
Keywords: batch effect; bone marrow; cell therapy; chondrogenesis; donor variation; human platelet lysate (hPL); regenerative medicine; stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
-
- Matsushita Y., Nagata M., Kozloff K.M., Welch J.D., Mizuhashi K., Tokavanich N., Hallett S.A., Link D.C., Nagasawa T., Ono W., et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat. Commun. 2020;11:332. doi: 10.1038/s41467-019-14029-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 20102-F2001080-FPR "Cancer Cluster II"/Land Salzburg
- WISS 2025 F 2000237-FIP "STEBS"/Land Salzburg
- GE2512/2-2 and CRC1444 (Sven Geissler)/German Research Foundation (DFG)
- 779293 (HIPGEN)/European Union Horizon 2020 Research and Innovation Program
- 733006 PACE (Dirk Strunk and Hans-Dieter Volk/European Union's Horizon 2020 research and innovation program: 733006 PACE
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

