Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment
- PMID: 35326413
- PMCID: PMC8946653
- DOI: 10.3390/cells11060962
Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment
Abstract
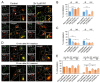
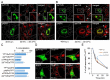
Phosphatidylinositol-3-phosphate (PI3P), a major identity tag of early endosomes (EEs), provides a platform for the recruitment of numerous cellular proteins containing an FYVE or PX domain that is required for PI3P-dependent maturation of EEs. Most of the PI3P in EEs is generated by the activity of Vps34, a catalytic component of class III phosphatidylinositol-3-phosphate kinase (PI3Ks) complex. In this study, we analyzed the role of Vps34-derived PI3P in the EE recycling circuit of unperturbed cells using VPS34-IN1 (IN1), a highly specific inhibitor of Vps34. IN1-mediated PI3P depletion resulted in the rapid dissociation of recombinant FYVE- and PX-containing PI3P-binding modules and endogenous PI3P-binding proteins, including EEA1 and EE sorting nexins. IN1 treatment triggered the rapid restructuring of EEs into a PI3P-independent functional configuration, and after IN1 washout, EEs were rapidly restored to a PI3P-dependent functional configuration. Analysis of the PI3P-independent configuration showed that the Vps34-derived PI3P is not essential for the pre-EE-associated functions and the fast recycling loop of the EE recycling circuit but contributes to EE maturation toward the degradation circuit, as previously shown in Vps34 knockout and knockdown studies. However, our study shows that Vps34-derived PI3P is also essential for the establishment of the Rab11a-dependent pathway, including recycling cargo sorting in this pathway and membrane flux from EEs to the pericentriolar endosomal recycling compartment (ERC). Rab11a endosomes of PI3P-depleted cells expanded and vacuolized outside the pericentriolar area without the acquisition of internalized transferrin (Tf). These endosomes had high levels of FIP5 and low levels of FIP3, suggesting that their maturation was arrested before the acquisition of FIP3. Consequently, Tf-loaded-, Rab11a/FIP5-, and Rab8a-positive endosomes disappeared from the pericentriolar area, implying that PI3P-associated functions are essential for ERC biogenesis. ERC loss was rapidly reversed after IN1 washout, which coincided with the restoration of FIP3 recruitment to Rab11a-positive endosomes and their dynein-dependent migration to the cell center. Thus, our study shows that Vps34-derived PI3P is indispensable in the recycling circuit to maintain the slow recycling pathway and biogenesis of the ERC.
Keywords: Rab11a endosomes; VPS34-IN1; Vps34; endosomal recycling compartment; phosphatidylinositol-3-phosphate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase.Biochem J. 2014 Nov 1;463(3):413-27. doi: 10.1042/BJ20140889. Biochem J. 2014. PMID: 25177796 Free PMC article.
-
Endosomal Phosphatidylinositol-3-Phosphate-Associated Functions Are Dispensable for Establishment of the Cytomegalovirus Pre-Assembly Compartment but Essential for the Virus Growth.Life (Basel). 2021 Aug 22;11(8):859. doi: 10.3390/life11080859. Life (Basel). 2021. PMID: 34440603 Free PMC article.
-
Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis.PLoS One. 2013 Oct 3;8(10):e76405. doi: 10.1371/journal.pone.0076405. eCollection 2013. PLoS One. 2013. PMID: 24098492 Free PMC article.
-
Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics.FEBS J. 2017 May;284(9):1267-1278. doi: 10.1111/febs.13987. Epub 2017 Jan 5. FEBS J. 2017. PMID: 27973739 Review.
-
How phosphoinositides shape autophagy in plant cells.Plant Sci. 2019 Apr;281:146-158. doi: 10.1016/j.plantsci.2019.01.017. Epub 2019 Feb 14. Plant Sci. 2019. PMID: 30824047 Review.
Cited by
-
An Overview of the Dry Eye Disease in Sjögren's Syndrome Using Our Current Molecular Understanding.Int J Mol Sci. 2023 Jan 13;24(2):1580. doi: 10.3390/ijms24021580. Int J Mol Sci. 2023. PMID: 36675090 Free PMC article. Review.
-
RUFY3 regulates endolysosomes perinuclear positioning, antigen presentation and migration in activated phagocytes.Nat Commun. 2023 Jul 18;14(1):4290. doi: 10.1038/s41467-023-40062-x. Nat Commun. 2023. PMID: 37463962 Free PMC article.
-
Contribution of Sorting Nexin 3 in the Cytomegalovirus Assembly.Biomedicines. 2025 Apr 11;13(4):936. doi: 10.3390/biomedicines13040936. Biomedicines. 2025. PMID: 40299528 Free PMC article.
-
PIKI-1, a class II phosphatidylinositol 3-kinase, functions in endocytic trafficking.bioRxiv [Preprint]. 2025 May 23:2025.05.22.655458. doi: 10.1101/2025.05.22.655458. bioRxiv. 2025. PMID: 40475523 Free PMC article. Preprint.
-
SNX27:Retromer:ESCPE-1-mediated early endosomal tubulation impacts cytomegalovirus replication.Front Cell Infect Microbiol. 2024 Sep 18;14:1399761. doi: 10.3389/fcimb.2024.1399761. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39359939 Free PMC article.
References
-
- Leonard D., Hayakawa A., Lawe D., Lambright D., Bellve K.D., Standley C., Lifshitz L.M., Fogarty K.E., Corvera S. Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J. Cell Sci. 2008;121:3445–3458. doi: 10.1242/jcs.031484. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

