Conversion of a Non-Cancer-Selective Promoter into a Cancer-Selective Promoter
- PMID: 35326649
- PMCID: PMC8946048
- DOI: 10.3390/cancers14061497
Conversion of a Non-Cancer-Selective Promoter into a Cancer-Selective Promoter
Abstract
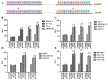
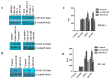
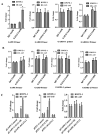
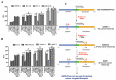
Progression-elevated gene-3 (PEG-3) and rat growth arrest and DNA damage-inducible gene-34 (GADD34) display significant sequence homology with regulation predominantly transcriptional. The rat full-length (FL) and minimal (min) PEG-3 promoter display cancer-selective expression in rodent and human tumors, allowing for cancer-directed regulation of transgenes, viral replication and in vivo imaging of tumors and metastases in animals, whereas the FL- and min-GADD34-Prom lack cancer specificity. Min-PEG-Prom and min-GADD34-Prom have identical sequences except for two single-point mutation differences (at -260 bp and +159 bp). Engineering double mutations in the min-GADD34-Prom produce the GAPE-Prom. Changing one base pair (+159) or both point mutations in the min-GADD34-Prom, but not the FL-GADD34-Prom, results in cancer-selective transgene expression in diverse cancer cells (including prostate, breast, pancreatic and neuroblastoma) vs. normal counterparts. Additionally, we identified a GATA2 transcription factor binding site, promoting cancer specificity when both min-PEG-Prom mutations are present in the GAPE-Prom. Taken together, introducing specific point mutations in a rat min-GADD34-Prom converts this non-cancer-specific promoter into a cancer-selective promoter, and the addition of GATA2 with existing AP1 and PEA3 transcription factors enhances further cancer-selective activity of the GAPE-Prom. The GAPE-Prom provides a genetic tool to specifically regulate transgene expression in cancer cells.
Keywords: GADD34; GATA2; PEG-3; cancer selective promoter; transcriptional regulation; tumor and metastasis imaging.
Conflict of interest statement
P.B.F. is a scientific co-founder and has equity in InterLeukin Combinatorial Therapies, Inc. (ILCT). VCU also has equity in ILCT. L.E. is the PI of a sponsored research agreement with ILCT, which is being managed by VCU. L.E. is also the PI of a Catalyst Matching Fund grant between ILCT and VCU. The other authors declare no conflict of interest.
Figures


Similar articles
-
Targeting gene expression selectively in cancer cells by using the progression-elevated gene-3 promoter.Proc Natl Acad Sci U S A. 2005 Jan 25;102(4):1059-64. doi: 10.1073/pnas.0409141102. Epub 2005 Jan 12. Proc Natl Acad Sci U S A. 2005. PMID: 15647352 Free PMC article.
-
Cooperation between AP1 and PEA3 sites within the progression elevated gene-3 (PEG-3) promoter regulate basal and differential expression of PEG-3 during progression of the oncogenic phenotype in transformed rat embryo cells.Oncogene. 2000 Jul 13;19(30):3411-21. doi: 10.1038/sj.onc.1203666. Oncogene. 2000. PMID: 10918598
-
Progression elevated gene-3 promoter (PEG-Prom) confers cancer cell selectivity to human polynucleotide phosphorylase (hPNPase(old-35))-mediated growth suppression.J Cell Physiol. 2008 May;215(2):401-9. doi: 10.1002/jcp.21320. J Cell Physiol. 2008. PMID: 17960560
-
Unique conditionally replication competent bipartite adenoviruses-cancer terminator viruses (CTV): efficacious reagents for cancer gene therapy.Cell Cycle. 2006 Jul;5(14):1531-6. doi: 10.4161/cc.5.14.3095. Epub 2006 Jul 17. Cell Cycle. 2006. PMID: 16861924 Review.
-
Mediterranean dietary traditions for the molecular treatment of human cancer: anti-oncogenic actions of the main olive oil's monounsaturated fatty acid oleic acid (18:1n-9).Curr Pharm Biotechnol. 2006 Dec;7(6):495-502. doi: 10.2174/138920106779116900. Curr Pharm Biotechnol. 2006. PMID: 17168666 Review.
References
-
- Chaffer C.L., Weinberg R.A. How does multistep tumorigenesis really proceed? Cancer Discov. 2015;5:22–24. doi: 10.1158/2159-8290.CD-14-0788. - DOI - PMC - PubMed
-
- Adolph K.W., Reddy P.G., Su Z.-Z., Fisher P.B. In: Chromosome and Genetic Analysis: Methods in Molecular Genetics. Adolph K.W., editor. Volume 1. Academic; Orlando, FL, USA: 1993. pp. 68–102.
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

