In Vitro, In Vivo, and In Silico Models of Lymphangiogenesis in Solid Malignancies
- PMID: 35326676
- PMCID: PMC8946816
- DOI: 10.3390/cancers14061525
In Vitro, In Vivo, and In Silico Models of Lymphangiogenesis in Solid Malignancies
Abstract
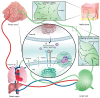
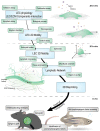
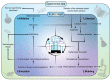
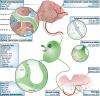
Lymphangiogenesis (LA) is the formation of new lymphatic vessels by lymphatic endothelial cells (LECs) sprouting from pre-existing lymphatic vessels. It is increasingly recognized as being involved in many diseases, such as in cancer and secondary lymphedema, which most often results from cancer treatments. For some cancers, excessive LA is associated with cancer progression and metastatic dissemination to the lymph nodes (LNs) through lymphatic vessels. The study of LA through in vitro, in vivo, and, more recently, in silico models is of paramount importance in providing novel insights and identifying the key molecular actors in the biological dysregulation of this process under pathological conditions. In this review, the different biological (in vitro and in vivo) models of LA, especially in a cancer context, are explained and discussed, highlighting their principal modeled features as well as their advantages and drawbacks. Imaging techniques of the lymphatics, complementary or even essential to in vivo models, are also clarified and allow the establishment of the link with computational approaches. In silico models are introduced, theoretically described, and illustrated with examples specific to the lymphatic system and the LA. Together, these models constitute a toolbox allowing the LA research to be brought to the next level.
Keywords: cancer; computational models; in silico methods; in vitro models; in vivo models; lymphangiogenesis; lymphatic endothelial cells; metastatic dissemination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Modeling lymphangiogenesis: Pairing in vitro and in vivo metrics.Microcirculation. 2023 Apr;30(2-3):e12802. doi: 10.1111/micc.12802. Epub 2023 Feb 28. Microcirculation. 2023. PMID: 36760223 Free PMC article. Review.
-
Lymphangiogenesis: in vitro and in vivo models.FASEB J. 2010 Jan;24(1):8-21. doi: 10.1096/fj.09-132852. Epub 2009 Sep 2. FASEB J. 2010. PMID: 19726757 Review.
-
The lymphatic vasculature: An active and dynamic player in cancer progression.Med Res Rev. 2022 Jan;42(1):576-614. doi: 10.1002/med.21855. Epub 2021 Sep 5. Med Res Rev. 2022. PMID: 34486138 Free PMC article. Review.
-
Podoplanin+ tumor lymphatics are rate limiting for breast cancer metastasis.PLoS Biol. 2018 Dec 28;16(12):e2005907. doi: 10.1371/journal.pbio.2005907. eCollection 2018 Dec. PLoS Biol. 2018. PMID: 30592710 Free PMC article.
-
Lymphatic endothelial cells, lymphedematous lymphangiogenesis, and molecular control of edema formation.Lymphat Res Biol. 2008;6(3-4):123-37. doi: 10.1089/lrb.2008.1005. Lymphat Res Biol. 2008. PMID: 19093784 Review.
References
-
- World Health Organization. [(accessed on 23 August 2021)]. Available online: https://www.who.int/health-topics/cancer#tab=overview.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

