pncCCND1_B Engages an Inhibitory Protein Network to Downregulate CCND1 Expression upon DNA Damage
- PMID: 35326688
- PMCID: PMC8946712
- DOI: 10.3390/cancers14061537
pncCCND1_B Engages an Inhibitory Protein Network to Downregulate CCND1 Expression upon DNA Damage
Abstract
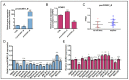
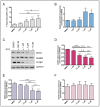
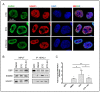
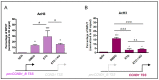
Promoter-associated noncoding RNAs (pancRNAs) represent a class of noncoding transcripts driven from the promoter region of protein-coding or non-coding genes that operate as cis-acting elements to regulate the expression of the host gene. PancRNAs act by altering the chromatin structure and recruiting transcription regulators. PncCCND1_B is driven by the promoter region of CCND1 and regulates CCND1 expression in Ewing sarcoma through recruitment of a multi-molecular complex composed of the RNA binding protein Sam68 and the DNA/RNA helicase DHX9. In this study, we investigated the regulation of CCND1 expression in Ewing sarcoma cells upon exposure to chemotherapeutic drugs. Pan-inhibitor screening indicated that etoposide, a drug used for Ewing sarcoma treatment, promotes transcription of pncCCND1_B and repression of CCND1 expression. RNA immunoprecipitation experiments showed increased binding of Sam68 to the pncCCND1_B after treatment, despite the significant reduction in DHX9 protein. This effect was associated with the formation of DNA:RNA duplexes at the CCND1 promoter. Furthermore, Sam68 interacted with HDAC1 in etoposide treated cells, thus contributing to chromatin remodeling and epigenetic changes. Interestingly, inhibition of the ATM signaling pathway by KU 55,933 treatment was sufficient to inhibit etoposide-induced Sam68-HDAC1 interaction without rescuing DHX9 expression. In these conditions, the DNA:RNA hybrids persist, thus contributing to the local chromatin inactivation at the CCND1 promoter region. Altogether, our results show an active role of Sam68 in DNA damage signaling and chromatin remodeling on the CCND1 gene by fine-tuning transitions of epigenetic complexes on the CCND1 promoter.
Keywords: CCND1; DNA damage; Ewing sarcoma; Sam68; noncoding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

