Antimicrobial Compounds from Microorganisms
- PMID: 35326749
- PMCID: PMC8944786
- DOI: 10.3390/antibiotics11030285
Antimicrobial Compounds from Microorganisms
Abstract
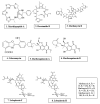
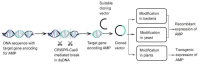
Antimicrobial resistance is an exigent public health concern owing to the emergence of novel strains of human resistant pathogens and the concurrent rise in multi-drug resistance. An influx of new antimicrobials is urgently required to improve the treatment outcomes of infectious diseases and save lives. Plant metabolites and bioactive compounds from chemical synthesis have found their efficacy to be dwindling, despite some of them being developed as drugs and used to treat human infections for several decades. Microorganisms are considered untapped reservoirs for promising biomolecules with varying structural and functional antimicrobial activity. The advent of cost-effective and convenient model organisms, state-of-the-art molecular biology, omics technology, and machine learning has enhanced the bioprospecting of novel antimicrobial drugs and the identification of new drug targets. This review summarizes antimicrobial compounds isolated from microorganisms and reports on the modern tools and strategies for exploiting promising antimicrobial drug candidates. The investigation identified a plethora of novel compounds from microbial sources with excellent antimicrobial activity against disease-causing human pathogens. Researchers could maximize the use of novel model systems and advanced biomolecular and computational tools in exploiting lead antimicrobials, consequently ameliorating antimicrobial resistance.
Keywords: antimicrobial peptides; drug discovery; microorganisms; model organisms; natural products; omics-informed drug discovery; secondary metabolites; structure-activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- O’Neill J. Antimicrobial Resistance:Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014
Publication types
LinkOut - more resources
Full Text Sources