Polymyxin Induces Significant Transcriptomic Perturbations of Cellular Signalling Networks in Human Lung Epithelial Cells
- PMID: 35326770
- PMCID: PMC8944768
- DOI: 10.3390/antibiotics11030307
Polymyxin Induces Significant Transcriptomic Perturbations of Cellular Signalling Networks in Human Lung Epithelial Cells
Abstract
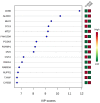
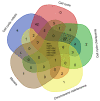
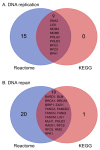
Inhaled polymyxins are increasingly used to treat pulmonary infections caused by multidrug-resistant Gram-negative pathogens. We have previously shown that apoptotic pathways, autophagy and oxidative stress are involved in polymyxin-induced toxicity in human lung epithelial cells. In the present study, we employed human lung epithelial cells A549 treated with polymyxin B as a model to elucidate the complex interplay of multiple signalling networks underpinning cellular responses to polymyxin toxicity. Polymyxin B induced toxicity (1.0 mM, 24 h) in A549 cells was assessed by flow cytometry and transcriptomics was performed using microarray. Polymyxin B induced cell death was 19.0 ± 4.2% at 24 h. Differentially expressed genes (DEGs) between the control and polymyxin B treated cells were identified with Student’s t-test. Pathway analysis was conducted with KEGG and Reactome and key hub genes related to polymyxin B induced toxicity were examined using the STRING database. In total we identified 899 DEGs (FDR < 0.01), KEGG and Reactome pathway analyses revealed significantly up-regulated genes related to cell cycle, DNA repair and DNA replication. NF-κB and nucleotide-binding oligomerization domain-like receptor (NOD) signalling pathways were identified as markedly down-regulated genes. Network analysis revealed the top 5 hub genes (i.e., degree) affected by polymyxin B treatment were PLK1(48), CDK20 (46), CCNA2 (42), BUB1 (40) and BUB1B (37). Overall, perturbations of cell cycle, DNA damage and pro-inflammatory NF-κB and NOD-like receptor signalling pathways play key roles in polymyxin-induced toxicity in human lung epithelial cells. Noting that NOD-like receptor signalling represents a group of key sensors for microorganisms and damage in the lung, understanding the mechanism of polymyxin-induced pulmonary toxicity will facilitate the optimisation of polymyxin inhalation therapy in patients.
Keywords: lung epithelial cells; polymyxin; pulmonary toxicity; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Transcriptomic Responses to Polymyxin B and Analogues in Human Kidney Tubular Cells.Antibiotics (Basel). 2023 Feb 20;12(2):415. doi: 10.3390/antibiotics12020415. Antibiotics (Basel). 2023. PMID: 36830325 Free PMC article.
-
Polymyxin-Induced Metabolic Perturbations in Human Lung Epithelial Cells.Antimicrob Agents Chemother. 2021 Aug 17;65(9):e0083521. doi: 10.1128/AAC.00835-21. Epub 2021 Aug 17. Antimicrob Agents Chemother. 2021. PMID: 34228550 Free PMC article.
-
Potential Toxicity of Polymyxins in Human Lung Epithelial Cells.Antimicrob Agents Chemother. 2017 May 24;61(6):e02690-16. doi: 10.1128/AAC.02690-16. Print 2017 Jun. Antimicrob Agents Chemother. 2017. PMID: 28416543 Free PMC article.
-
Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review.J Antimicrob Chemother. 2007 Dec;60(6):1206-15. doi: 10.1093/jac/dkm357. Epub 2007 Sep 17. J Antimicrob Chemother. 2007. PMID: 17878146 Review.
-
Clinical use of intravenous polymyxin B for the treatment of patients with multidrug-resistant Gram-negative bacterial infections: An evaluation of the current evidence.J Glob Antimicrob Resist. 2021 Mar;24:342-359. doi: 10.1016/j.jgar.2020.12.026. Epub 2021 Jan 21. J Glob Antimicrob Resist. 2021. PMID: 33486122 Review.
Cited by
-
Antimicrobial Peptides towards Clinical Application-A Long History to Be Concluded.Int J Mol Sci. 2024 Apr 29;25(9):4870. doi: 10.3390/ijms25094870. Int J Mol Sci. 2024. PMID: 38732089 Free PMC article. Review.
References
-
- World Health Organization Ten Threats to Global Health in 2019. [(accessed on 20 February 2022)]. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-....
-
- Naghavi M., Abajobir A.A., Abbafati C., Abbas K.M., Abd-Allah F., Abera S.F., Aboyans V., Adetokunboh O., Afshin A., Agrawal A., et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. - DOI - PMC - PubMed
-
- Troeger C., Blacker B., Khalil I.A., Rao P.C., Cao J., Zimsen S.R.M., Albertson S.B., Deshpande A., Farag T., Abebe Z., et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018;18:1191–1210. doi: 10.1016/S1473-3099(18)30310-4. - DOI - PMC - PubMed
-
- Castanheira M., Deshpande L.M., Mendes R.E., Canton R., Sader H.S., Jones R.N. Variations in the occurrence of resistance phenotypes and carbapenemase genes among Enterobacteriaceae isolates in 20 years of the SENTRY Antimicrobial Surveillance Program. Open Forum Infect. Dis. 2019;6((Suppl. 1)):S23–S33. doi: 10.1093/ofid/ofy347. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

