RecA and Specialized Error-Prone DNA Polymerases Are Not Required for Mutagenesis and Antibiotic Resistance Induced by Fluoroquinolones in Pseudomonas aeruginosa
- PMID: 35326787
- PMCID: PMC8944484
- DOI: 10.3390/antibiotics11030325
RecA and Specialized Error-Prone DNA Polymerases Are Not Required for Mutagenesis and Antibiotic Resistance Induced by Fluoroquinolones in Pseudomonas aeruginosa
Abstract
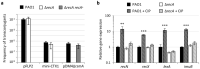
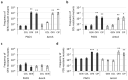
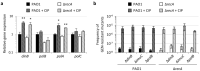
To cope with stressful conditions, including antibiotic exposure, bacteria activate the SOS response, a pathway that induces error-prone DNA repair and mutagenesis mechanisms. In most bacteria, the SOS response relies on the transcriptional repressor LexA and the co-protease RecA, the latter being also involved in homologous recombination. The role of the SOS response in stress- and antibiotic-induced mutagenesis has been characterized in detail in the model organism Escherichia coli. However, its effect on antibiotic resistance in the human pathogen Pseudomonas aeruginosa is less clear. Here, we analyzed a recA deletion mutant and confirmed, by conjugation and gene expression assays, that RecA is required for homologous recombination and SOS response induction in P. aeruginosa. MIC assays demonstrated that RecA affects P. aeruginosa resistance only towards fluoroquinolones and genotoxic agents. The comparison of antibiotic-resistant mutant frequency between treated and untreated cultures revealed that, among the antibiotics tested, only fluoroquinolones induced mutagenesis in P. aeruginosa. Notably, both RecA and error-prone DNA polymerases were found to be dispensable for this process. These data demonstrate that the SOS response is not required for antibiotic-induced mutagenesis in P. aeruginosa, suggesting that RecA inhibition is not a suitable strategy to target antibiotic-induced emergence of resistance in this pathogen.
Keywords: DinB; ImuBC; PolB; SOS response; homologous recombination; induced mutagenesis; resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
DinB (DNA polymerase IV), ImuBC and RpoS contribute to the generation of ciprofloxacin-resistance mutations in Pseudomonas aeruginosa.Mutat Res. 2023 Jul-Dec;827:111836. doi: 10.1016/j.mrfmmm.2023.111836. Epub 2023 Aug 12. Mutat Res. 2023. PMID: 37625357
-
Systematically Altering Bacterial SOS Activity under Stress Reveals Therapeutic Strategies for Potentiating Antibiotics.mSphere. 2016 Aug 10;1(4):e00163-16. doi: 10.1128/mSphere.00163-16. eCollection 2016 Jul-Aug. mSphere. 2016. PMID: 27536734 Free PMC article.
-
ImuB and ImuC contribute to UV-induced mutagenesis as part of the SOS regulon in Pseudomonas aeruginosa.Environ Mol Mutagen. 2019 Aug;60(7):594-601. doi: 10.1002/em.22290. Epub 2019 Apr 30. Environ Mol Mutagen. 2019. PMID: 30921487
-
Coexistence of SOS-Dependent and SOS-Independent Regulation of DNA Repair Genes in Radiation-Resistant Deinococcus Bacteria.Cells. 2021 Apr 16;10(4):924. doi: 10.3390/cells10040924. Cells. 2021. PMID: 33923690 Free PMC article. Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
The diverse phenotypic and mutational landscape induced by fluoroquinolone treatment.mSystems. 2025 Aug 19;10(8):e0071325. doi: 10.1128/msystems.00713-25. Epub 2025 Jul 31. mSystems. 2025. PMID: 40742148 Free PMC article.
-
A role for the stringent response in ciprofloxacin resistance in Pseudomonas aeruginosa.Sci Rep. 2024 Apr 13;14(1):8598. doi: 10.1038/s41598-024-59188-z. Sci Rep. 2024. PMID: 38615146 Free PMC article.
-
Metabolic and transcriptional activities underlie stationary-phase Pseudomonas aeruginosa sensitivity to Levofloxacin.Microbiol Spectr. 2024 Jan 11;12(1):e0356723. doi: 10.1128/spectrum.03567-23. Epub 2023 Dec 11. Microbiol Spectr. 2024. PMID: 38078717 Free PMC article.
-
Fast evolution of SOS-independent multi-drug resistance in bacteria.Elife. 2025 Jul 9;13:RP95058. doi: 10.7554/eLife.95058. Elife. 2025. PMID: 40631979 Free PMC article.
-
The histidine kinase NahK regulates denitrification and nitric oxide accumulation through RsmA in Pseudomonas aeruginosa.J Bacteriol. 2025 Jan 31;207(1):e0040824. doi: 10.1128/jb.00408-24. Epub 2024 Dec 11. J Bacteriol. 2025. PMID: 39660891 Free PMC article.
References
Grants and funding
- PRIN 2017 - 20177J5Y3P/Italian Ministry of Education, University and Research
- PRIN 2020 - 20208LLXEJ/Italian Ministry of Education, University and Research
- Excellence Departments (Art. 1, commi 314-337 Legge 232/2016)/Italian Ministry of Education, University and Research
- Gruppi di Ricerca 2020 - POR A0375E0026/Regione Lazio
LinkOut - more resources
Full Text Sources

