New Glycosylated Polyene Macrolides: Refining the Ore from Genome Mining
- PMID: 35326797
- PMCID: PMC8944477
- DOI: 10.3390/antibiotics11030334
New Glycosylated Polyene Macrolides: Refining the Ore from Genome Mining
Abstract
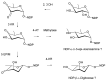
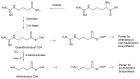
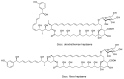
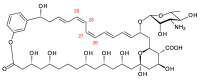
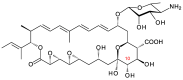
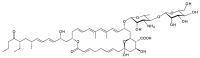
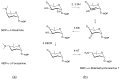
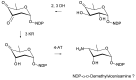
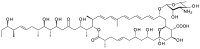
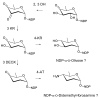
Glycosylated polyene macrolides include effective antifungal agents, such as pimaricin, nystatin, candicidin, and amphotericin B. For the treatment of systemic mycoses, amphotericin B has been described as a gold-standard antibiotic because of its potent activity against a broad spectrum of fungal pathogens, which do not readily become resistant. However, amphotericin B has severe toxic side effects, and the development of safer alternatives remains an important objective. One approach towards obtaining such compounds is to discover new related natural products. Advances in next-generation sequencing have delivered a wealth of microbial genome sequences containing polyene biosynthetic gene clusters. These typically encode a modular polyketide synthase that catalyzes the assembly of the aglycone core, a cytochrome P450 that oxidizes a methyl branch to a carboxyl group, and additional enzymes for synthesis and attachment of a single mycosamine sugar residue. In some cases, further P450s catalyze epoxide formation or hydroxylation within the macrolactone. Bioinformatic analyses have identified over 250 of these clusters. Some are predicted to encode potentially valuable new polyenes that have not been uncovered by traditional screening methods. Recent experimental studies have characterized polyenes with new polyketide backbones, previously unknown late oxygenations, and additional sugar residues that increase water-solubility and reduce hemolytic activity. Here we review these studies and assess how this new knowledge can help to prioritize silent polyene clusters for further investigation. This approach should improve the chances of discovering better antifungal antibiotics.
Keywords: antifungal antibiotics; biosynthetic gene clusters; genome mining; glycosylated polyene macrolide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







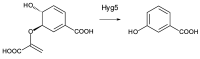





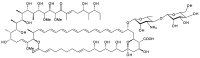







Similar articles
-
Polyene antibiotic biosynthesis gene clusters.Appl Microbiol Biotechnol. 2003 May;61(3):179-88. doi: 10.1007/s00253-002-1183-5. Epub 2002 Dec 18. Appl Microbiol Biotechnol. 2003. PMID: 12698274 Review.
-
Biosynthetic engineering of polyene macrolides towards generation of improved antifungal and antiparasitic agents.Curr Top Med Chem. 2008;8(8):639-53. doi: 10.2174/156802608784221479. Curr Top Med Chem. 2008. PMID: 18473889 Review.
-
Polyene macrolide biosynthesis in streptomycetes and related bacteria: recent advances from genome sequencing and experimental studies.Appl Microbiol Biotechnol. 2016 May;100(9):3893-908. doi: 10.1007/s00253-016-7474-z. Epub 2016 Mar 29. Appl Microbiol Biotechnol. 2016. PMID: 27023916 Review.
-
Nystatin-like Pseudonocardia polyene B1, a novel disaccharide-containing antifungal heptaene antibiotic.Sci Rep. 2018 Sep 11;8(1):13584. doi: 10.1038/s41598-018-31801-y. Sci Rep. 2018. PMID: 30206268 Free PMC article.
-
New nystatin-related antifungal polyene macrolides with altered polyol region generated via biosynthetic engineering of Streptomyces noursei.Appl Environ Microbiol. 2011 Sep;77(18):6636-43. doi: 10.1128/AEM.05780-11. Epub 2011 Jul 15. Appl Environ Microbiol. 2011. PMID: 21764946 Free PMC article.
Cited by
-
Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855.Int J Mol Sci. 2024 Nov 27;25(23):12744. doi: 10.3390/ijms252312744. Int J Mol Sci. 2024. PMID: 39684453 Free PMC article.
-
A polyene macrolide targeting phospholipids in the fungal cell membrane.Nature. 2025 Apr;640(8059):743-751. doi: 10.1038/s41586-025-08678-9. Epub 2025 Mar 19. Nature. 2025. PMID: 40108452 Free PMC article.
-
Polyene Antibiotics Physical Chemistry and Their Effect on Lipid Membranes; Impacting Biological Processes and Medical Applications.Membranes (Basel). 2022 Jun 30;12(7):681. doi: 10.3390/membranes12070681. Membranes (Basel). 2022. PMID: 35877884 Free PMC article. Review.
-
Novel Tetraene Macrodiolides Are Effective Inducers of Mitochondrial Apoptosis in Jurkat Cells.Int J Mol Sci. 2025 May 27;26(11):5139. doi: 10.3390/ijms26115139. Int J Mol Sci. 2025. PMID: 40507950 Free PMC article.
-
BAC cloning and heterologous expression of a giant biosynthetic gene cluster encoding antifungal neotetrafibricin in streptomyces rubrisoli.Front Bioeng Biotechnol. 2022 Aug 15;10:964765. doi: 10.3389/fbioe.2022.964765. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36046673 Free PMC article.
References
-
- Omura S., Tanaka H. Production, Structure and Antifungal Activity of Polyene Macrolides. In: Omura S., editor. Macrolide Antibiotics, Chemistry, Biology and Practice. Academic Press; New York, NY, USA: 1986. pp. 351–404.
-
- Abu-Salah K.M. Amphotericin B: An update. Br. J. Biomed. Sci. 1996;53:8757689. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

