Development and Application of a High-Resolution Melting Analysis with Unlabeled Probes for the Screening of Short-Tailed Sheep TBXT Heterozygotes
- PMID: 35327188
- PMCID: PMC8944613
- DOI: 10.3390/ani12060792
Development and Application of a High-Resolution Melting Analysis with Unlabeled Probes for the Screening of Short-Tailed Sheep TBXT Heterozygotes
Abstract
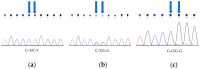
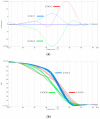
The short-tailed phenotype has long been considered one of the best traits for population genetic improvement in sheep breeding. In short-tailed sheep, not only is tail fat eliminated but also the pubic area is exposed due to the lack of a tail covering, giving them an advantage in reproduction. Recent studies have shown that two linked mutations in sheep TBXT at nucleotides 333 and 334 are associated with the short-tailed phenotype. In the population of short-tailed sheep, several heterozygous mutants of this gene are found. In our research, we used high-resolution melting (HRM) to identify homozygous and heterozygous genotypes in a flock of short-tailed sheep and compared the results with those of Sanger sequencing, which were identical. This demonstrates that our established HRM method, a rapid and inexpensive genotyping method, can be used to identify homozygous and heterozygous individuals in short-tailed sheep flocks.
Keywords: HRM; TBXT; sheep; short-tailed trait.
Conflict of interest statement
G.C., G.Y., Y.S., A.D., T.M., C.W., H.S., M.L., D.W., L.C., X.X., T.H., Y.Y., J.S. and X.L. are co-authors of an issued Chinese invention patent (as described above) covering the high-resolution melting curve used in this study. The efficacy of this patented approach has not been demonstrated nor do the authors believe the patent or any future benefits they may receive because of it inappropriately influence any work described in this manuscript. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Association of Sheep Tail Type with the T Gene Single Nucleotide Polymorphisms Loci.Life (Basel). 2025 Feb 21;15(3):342. doi: 10.3390/life15030342. Life (Basel). 2025. PMID: 40141686 Free PMC article.
-
Exploring the formation mechanism of short-tailed phenotypes in animals using mutant mice with the TBXT gene c.G334T developed by CRISPR/Cas9.Gene. 2024 Jun 5;910:148310. doi: 10.1016/j.gene.2024.148310. Epub 2024 Feb 23. Gene. 2024. PMID: 38401832
-
Two linked TBXT (brachyury) gene polymorphisms are associated with the tailless phenotype in fat-rumped sheep.Anim Genet. 2019 Dec;50(6):772-777. doi: 10.1111/age.12852. Epub 2019 Sep 2. Anim Genet. 2019. PMID: 31475743 Free PMC article.
-
High Resolution Melting as a rapid, reliable, accurate and cost-effective emerging tool for genotyping pathogenic bacteria and enhancing molecular epidemiological surveillance: a comprehensive review of the literature.Ann Ig. 2017 Jul-Aug;29(4):293-316. doi: 10.7416/ai.2017.2153. Ann Ig. 2017. PMID: 28569339 Review.
-
Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways.Anim Genet. 2021 Dec;52(6):799-812. doi: 10.1111/age.13133. Epub 2021 Sep 1. Anim Genet. 2021. PMID: 34472112 Review.
Cited by
-
Association of Sheep Tail Type with the T Gene Single Nucleotide Polymorphisms Loci.Life (Basel). 2025 Feb 21;15(3):342. doi: 10.3390/life15030342. Life (Basel). 2025. PMID: 40141686 Free PMC article.
References
-
- Hickman G.C. The mammalian tail: A review of functions. Mammal Rev. 1979;9:143–157. doi: 10.1111/j.1365-2907.1979.tb00252.x. - DOI
-
- Zhu C., Cheng H., Li N., Liu T., Ma Y. Isobaric tags for relative and absolute quantification-based proteomics reveals candidate proteins of fat deposition in chinese indigenous sheep with morphologically different tails. Front. Genet. 2021;12:710449. doi: 10.3389/fgene.2021.710449. - DOI - PMC - PubMed
-
- Abied A., Bagadi A., Bordbar F., Pu Y., Augustino S.M., Xue X., Xing F., Gebreselassie G., Han J.-L., Mwacharo J.-L.H.J.M., et al. Genomic diversity, population structure, and signature of selection in five chinese native sheep breeds adapted to extreme environments. Genes. 2020;11:494. doi: 10.3390/genes11050494. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

