Differential Transcriptome Profiling Unveils Novel Deregulated Gene Signatures Involved in Pathogenesis of Alzheimer's Disease
- PMID: 35327413
- PMCID: PMC8945049
- DOI: 10.3390/biomedicines10030611
Differential Transcriptome Profiling Unveils Novel Deregulated Gene Signatures Involved in Pathogenesis of Alzheimer's Disease
Abstract
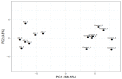
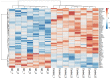
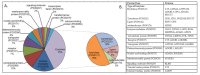
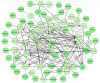
Alzheimer’s disease (AD) is a neurodegenerative disorder that is characterized by a progressive loss of cognitive functions at a higher level than normal aging. Although the apolipoprotein (APOE) gene is a major risk factor in developing AD, other genes have also been reported to be linked with complex phenotypes. Therefore, this genome-wide expression study explored differentially expressed genes as possible novel biomarkers involved in AD. The mRNA expression dataset, GSE28146, containing 15 sample data composed of 7 AD cases from the hippocampus region with age-matched control (n = 8, >80 years), was analyzed. Using “affy” R-package, mRNA expression was calculated, while pathway enrichment analysis was performed to determine related biological processes. Of 58 differentially expressed genes, 44 downregulated and 14 upregulated genes were found to be significantly (p < 0.001) altered. The pathway enrichment analysis revealed two altered genes, i.e., dynein light chain 1 (DYNLL1) and kalirin (KLRN), associated with AD in the elderly population. The majority of genes were associated with retrograde endocannabinoid as well as vascular endothelial growth factors affecting the complex phenotypes. The DYNLL1 and KLRN genes may be involved with AD and Huntington’s disease (HD) phenotypes and represent a common genetic basis of these diseases. However, the hallmark of AD is dementia, while the classic motor sign of HD includes chorea. Our data warrant further investigation to identify the role of these genes in disease pathogenesis.
Keywords: Alzheimer’s disease; differentially expressed genes; microarray analysis; transcriptome analysis.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

