Oral Microbiome: Getting to Know and Befriend Neighbors, a Biological Approach
- PMID: 35327473
- PMCID: PMC8945538
- DOI: 10.3390/biomedicines10030671
Oral Microbiome: Getting to Know and Befriend Neighbors, a Biological Approach
Abstract
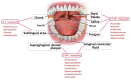
The oral microbiome, forming a biofilm that covers the oral structures, contains a high number of microorganisms. Biofilm formation starts from the salivary pellicle that allows bacterial adhesion-colonization-proliferation, co-aggregation and biofilm maturation in a complex microbial community. There is a constant bidirectional crosstalk between human host and its oral microbiome. The paper presents the fundamentals regarding the oral microbiome and its relationship to modulator factors, oral and systemic health. The modern studies of oral microorganisms and relationships with the host benefits are based on genomics, transcriptomics, proteomics and metabolomics. Pharmaceuticals such as antimicrobials, prebiotics, probiotics, surface active or abrasive agents and plant-derived ingredients may influence the oral microbiome. Many studies found associations between oral dysbiosis and systemic disorders, including autoimmune diseases, cardiovascular, diabetes, cancers and neurodegenerative disorders. We outline the general and individual factors influencing the host-microbial balance and the possibility to use the analysis of the oral microbiome in prevention, diagnosis and treatment in personalized medicine. Future therapies should take in account the restoration of the normal symbiotic relation with the oral microbiome.
Keywords: Oral Microbiome Database; amyloid; autoimmune diseases; biofilm; genomics; immune responses; metabolomics; oral diseases; systemic diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Margulis L. Symbiosis in Cell Evolution. 2nd ed. W.H. Freeman; New York, NY, USA: 1993. p. 452.
-
- Gordon J., Knowlton N., Relman D.A., Rohwer F., Youle M. Superorganisms and holobionts. Microbe. 2013;8:152–153. doi: 10.1128/microbe.8.152.1. - DOI
Publication types
LinkOut - more resources
Full Text Sources

