Proteomic Profiling and T Cell Receptor Usage of Abacavir Susceptible Subjects
- PMID: 35327495
- PMCID: PMC8945713
- DOI: 10.3390/biomedicines10030693
Proteomic Profiling and T Cell Receptor Usage of Abacavir Susceptible Subjects
Abstract
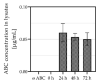
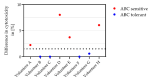
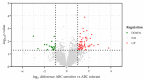
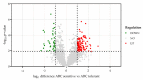
Type B adverse drug reactions (ADRs) represent a significant threat as their occurrence arises unpredictable and despite proper application of the drug. The severe immune reaction Abacavir Hypersensitivity Syndrome (AHS) that arises in HIV+ patients treated with the antiretroviral drug Abacavir (ABC) strongly correlates to the presence of the human leukocyte antigen (HLA) genotype HLA-B*57:01 and discriminates HLA-B*57:01+ HIV+ patients from ABC treatment. However, not all HLA-B*57:01+ HIV+ patients are affected by AHS, implying the involvement of further patient-specific factors in the development of AHS. The establishment of a reliable assay to classify HLA-B*57:01 carriers as ABC sensitive or ABC tolerant allowed to investigate the T cell receptor (TCR) Vβ chain repertoire of effector cells and revealed Vβ6 and Vβ24 as potential public TCRs in ABC sensitive HLA-B*57:01 carriers. Furthermore, distinct effects of ABC on the cellular proteome of ABC sensitive and tolerant volunteers were observed and suggest enhanced activation and maturation of dentritic cells (DC) in ABC sensitive volunteers. Analysis of ABC-naïve cellular proteomes identified the T cell immune regulator 1 (TCIRG1) as a potential prognostic biomarker for ABC susceptibility and the involvement of significantly upregulated proteins, particularly in peptide processing, antigen presentation, interferon (IFN), and cytokine regulation.
Keywords: HLA-B*57:01; T cell receptor; abacavir; adverse drug reaction; hypersensitivity; proteome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Unravelling the Proteomics of HLA-B*57:01+ Antigen Presenting Cells during Abacavir Medication.J Pers Med. 2022 Jan 4;12(1):40. doi: 10.3390/jpm12010040. J Pers Med. 2022. PMID: 35055355 Free PMC article.
-
New genetic predictors for abacavir tolerance in HLA-B*57:01 positive individuals.Hum Immunol. 2020 Jun;81(6):300-304. doi: 10.1016/j.humimm.2020.02.011. Epub 2020 Mar 12. Hum Immunol. 2020. PMID: 32173028 Free PMC article.
-
TARC/CCL17 Expression Is Associated with CD8+ T Cell Recruitment in Abacavir-Induced Skin Hypersensitivity in HLA-Transgenic Mice.Biol Pharm Bull. 2022;45(9):1347-1353. doi: 10.1248/bpb.b22-00313. Biol Pharm Bull. 2022. PMID: 36047204
-
Successful translation of pharmacogenetics into the clinic: the abacavir example.Mol Diagn Ther. 2009;13(1):1-9. doi: 10.1007/BF03256308. Mol Diagn Ther. 2009. PMID: 19351209 Review.
-
Fever, rash, and systemic symptoms: understanding the role of virus and HLA in severe cutaneous drug allergy.J Allergy Clin Immunol Pract. 2014 Jan-Feb;2(1):21-33. doi: 10.1016/j.jaip.2013.11.005. J Allergy Clin Immunol Pract. 2014. PMID: 24565765 Free PMC article. Review.
Cited by
-
HLA-B*57:01/Carbamazepine-10,11-Epoxide Association Triggers Upregulation of the NFκB and JAK/STAT Pathways.Cells. 2023 Feb 21;12(5):676. doi: 10.3390/cells12050676. Cells. 2023. PMID: 36899812 Free PMC article.
References
-
- WHO International drug monitoring: The role of national centres. Report of a WHO meeting. World Health Organ. Tech. Rep. Ser. 1972;498:1–25. - PubMed
-
- Simper G.S., Celik A.A., Kunze-Schumacher H., Blasczyk R., Bade-Doding C. Physiology and Pathology of Drug Hypersensitivity: Role of Human Leukocyte Antigens. IntechOpen; London, UK: 2017.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

