The Role of Lipids in CRAC Channel Function
- PMID: 35327543
- PMCID: PMC8944985
- DOI: 10.3390/biom12030352
The Role of Lipids in CRAC Channel Function
Abstract
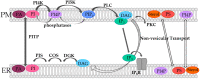
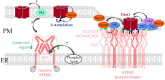
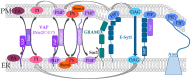
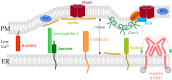
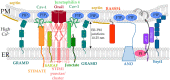
The composition and dynamics of the lipid membrane define the physical properties of the bilayer and consequently affect the function of the incorporated membrane transporters, which also applies for the prominent Ca2+ release-activated Ca2+ ion channel (CRAC). This channel is activated by receptor-induced Ca2+ store depletion of the endoplasmic reticulum (ER) and consists of two transmembrane proteins, STIM1 and Orai1. STIM1 is anchored in the ER membrane and senses changes in the ER luminal Ca2+ concentration. Orai1 is the Ca2+-selective, pore-forming CRAC channel component located in the plasma membrane (PM). Ca2+ store-depletion of the ER triggers activation of STIM1 proteins, which subsequently leads to a conformational change and oligomerization of STIM1 and its coupling to as well as activation of Orai1 channels at the ER-PM contact sites. Although STIM1 and Orai1 are sufficient for CRAC channel activation, their efficient activation and deactivation is fine-tuned by a variety of lipids and lipid- and/or ER-PM junction-dependent accessory proteins. The underlying mechanisms for lipid-mediated CRAC channel modulation as well as the still open questions, are presented in this review.
Keywords: CRAC channel; ER-PM junctions; Orai1; STIM1; lipids; modulatory proteins; protein-lipid interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation.Nature. 2008 Jul 24;454(7203):538-42. doi: 10.1038/nature07065. Epub 2008 Jul 2. Nature. 2008. PMID: 18596693 Free PMC article.
-
Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1).Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):13299-304. doi: 10.1073/pnas.1101664108. Epub 2011 Jul 25. Proc Natl Acad Sci U S A. 2011. PMID: 21788510 Free PMC article.
-
Defects in the STIM1 SOARα2 domain affect multiple steps in the CRAC channel activation cascade.Cell Mol Life Sci. 2021 Oct;78(19-20):6645-6667. doi: 10.1007/s00018-021-03933-4. Epub 2021 Sep 8. Cell Mol Life Sci. 2021. PMID: 34498097 Free PMC article.
-
More Than Just Simple Interaction between STIM and Orai Proteins: CRAC Channel Function Enabled by a Network of Interactions with Regulatory Proteins.Int J Mol Sci. 2021 Jan 5;22(1):471. doi: 10.3390/ijms22010471. Int J Mol Sci. 2021. PMID: 33466526 Free PMC article. Review.
-
Modulation of Orai1 and STIM1 by Cellular Factors.In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 4. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 4. PMID: 30299655 Free Books & Documents. Review.
Cited by
-
The Molecular Biology of Placental Transport of Calcium to the Human Foetus.Int J Mol Sci. 2025 Jan 4;26(1):383. doi: 10.3390/ijms26010383. Int J Mol Sci. 2025. PMID: 39796238 Free PMC article. Review.
-
CRAC and SK Channels: Their Molecular Mechanisms Associated with Cancer Cell Development.Cancers (Basel). 2022 Dec 23;15(1):101. doi: 10.3390/cancers15010101. Cancers (Basel). 2022. PMID: 36612099 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

