The Novel Protease Activities of JMJD5-JMJD6-JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled
- PMID: 35327545
- PMCID: PMC8945206
- DOI: 10.3390/biom12030347
The Novel Protease Activities of JMJD5-JMJD6-JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled
Abstract
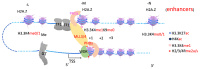
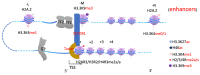
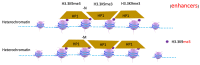
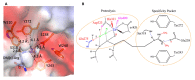
The surreptitious discoveries of the protease activities on arginine-methylated targets of a subfamily of Jumonji domain-containing family including JMJD5, JMJD6, and JMJD7 pose several questions regarding their authenticity, function, purpose, and relations with others. At the same time, despite several decades of efforts and massive accumulating data regarding the roles of the arginine methyltransferase family (PRMTs), the exact function of this protein family still remains a mystery, though it seems to play critical roles in transcription regulation, including activation and inactivation of a large group of genes, as well as other biological activities. In this review, we aim to elucidate that the function of JMJD5/6/7 and PRMTs are likely coupled. Besides roles in the regulation of the biogenesis of membrane-less organelles in cells, they are major players in regulating stimulating transcription factors to control the activities of RNA Polymerase II in higher eukaryotes, especially in the animal kingdom. Furthermore, we propose that arginine methylation by PRMTs could be a ubiquitous action marked for destruction after missions by a subfamily of the Jumonji protein family.
Keywords: JMJD5; JMJD6; JMJD7; Jumonji; PRMT.
Conflict of interest statement
G.Z. has ownership in NB Life Laboratory LLC.
Figures



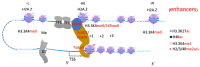


Similar articles
-
Clipping of arginine-methylated histone tails by JMJD5 and JMJD7.Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):E7717-E7726. doi: 10.1073/pnas.1706831114. Epub 2017 Aug 28. Proc Natl Acad Sci U S A. 2017. PMID: 28847961 Free PMC article.
-
Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.Sci Rep. 2018 Feb 19;8(1):3275. doi: 10.1038/s41598-018-21432-8. Sci Rep. 2018. PMID: 29459673 Free PMC article.
-
Elucidating the role of PRMTs in prostate cancer using open access databases and a patient cohort dataset.Histol Histopathol. 2023 Mar;38(3):287-302. doi: 10.14670/HH-18-513. Epub 2022 Sep 9. Histol Histopathol. 2023. PMID: 36082942
-
Protein arginine methylation/demethylation and cancer.Oncotarget. 2016 Oct 11;7(41):67532-67550. doi: 10.18632/oncotarget.11376. Oncotarget. 2016. PMID: 27556302 Free PMC article. Review.
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System.Mol Neurobiol. 2023 Oct;60(10):6060-6091. doi: 10.1007/s12035-023-03465-x. Epub 2023 Jul 6. Mol Neurobiol. 2023. PMID: 37415067 Review.
Cited by
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
-
Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.Sci Rep. 2022 Nov 30;12(1):20680. doi: 10.1038/s41598-022-24154-0. Sci Rep. 2022. PMID: 36450832 Free PMC article.
-
JMJD family proteins in cancer and inflammation.Signal Transduct Target Ther. 2022 Sep 1;7(1):304. doi: 10.1038/s41392-022-01145-1. Signal Transduct Target Ther. 2022. PMID: 36050314 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

