Characterization of a Programmable Argonaute Nuclease from the Mesophilic Bacterium Rummeliibacillus suwonensis
- PMID: 35327547
- PMCID: PMC8945025
- DOI: 10.3390/biom12030355
Characterization of a Programmable Argonaute Nuclease from the Mesophilic Bacterium Rummeliibacillus suwonensis
Abstract
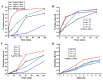
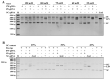
Prokaryotic Argonautes (pAgos) from mesophilic bacteria are attracting increasing attention for their genome editing potential. So far, it has been reported that KmAgo from Kurthia massiliensis can utilize DNA and RNA guide of any sequence to effectively cleave DNA and RNA targets. Here we find that three active pAgos, which have about 50% sequence identity with KmAgo, possess typical DNA-guided DNA target cleavage ability. Among them, RsuAgo from Rummeliibacillus suwonensis is mainly explored for which can cleave both DNA and RNA targets. Interestingly, RsuAgo-mediated RNA target cleavage occurs only with short guide DNAs in a narrow length range (16-20 nt), and mismatches between the guide and target sequence greatly affect the efficiency of RNA target cleavage. RsuAgo-mediated target cleavage shows a preference for a guide strand with a 5'-terminal A residue. Furthermore, we have found that RsuAgo can cleave double-stranded DNA in a low-salt buffer at 37 °C. These properties of RsuAgo provide a new tool for DNA and RNA manipulation at moderate temperatures.
Keywords: DNA cleavage; RNA cleavage; argonaute; mesophilic; nuclease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
A programmable pAgo nuclease with RNA target-cleavage specificity from the mesophilic bacterium Verrucomicrobia.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 10;55(8):1204-1212. doi: 10.3724/abbs.2023110. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37431184 Free PMC article.
-
A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis.Nucleic Acids Res. 2021 Apr 19;49(7):4054-4065. doi: 10.1093/nar/gkab182. Nucleic Acids Res. 2021. PMID: 33744962 Free PMC article.
-
A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis.Nucleic Acids Res. 2021 Feb 22;49(3):1597-1608. doi: 10.1093/nar/gkaa1278. Nucleic Acids Res. 2021. PMID: 33444443 Free PMC article.
-
A long look at short prokaryotic Argonautes.Trends Cell Biol. 2023 Jul;33(7):605-618. doi: 10.1016/j.tcb.2022.10.005. Epub 2022 Nov 22. Trends Cell Biol. 2023. PMID: 36428175 Review.
-
The evolutionary journey of Argonaute proteins.Nat Struct Mol Biol. 2014 Sep;21(9):743-53. doi: 10.1038/nsmb.2879. Nat Struct Mol Biol. 2014. PMID: 25192263 Free PMC article. Review.
Cited by
-
A Programmable, DNA-Exclusively-Guided Argonaute DNase and Its Higher Cleavage Specificity Achieved by 5'-Hydroxylated Guide.Biomolecules. 2022 Sep 21;12(10):1340. doi: 10.3390/biom12101340. Biomolecules. 2022. PMID: 36291549 Free PMC article.
-
Characterization of argonaute nucleases from mesophilic bacteria Pseudobutyrivibrio ruminis.Bioresour Bioprocess. 2024 Oct 7;11(1):94. doi: 10.1186/s40643-024-00797-x. Bioresour Bioprocess. 2024. PMID: 39373873 Free PMC article.
-
Mn2+-induced structural flexibility enhances the entire catalytic cycle and the cleavage of mismatches in prokaryotic argonaute proteins.Chem Sci. 2024 Mar 14;15(15):5612-5626. doi: 10.1039/d3sc06221j. eCollection 2024 Apr 17. Chem Sci. 2024. PMID: 38638240 Free PMC article.
-
Characterization of Argonaute Nuclease from Mesophilic Bacterium Chroococcidiopsis.Int J Mol Sci. 2025 Jan 27;26(3):1085. doi: 10.3390/ijms26031085. Int J Mol Sci. 2025. PMID: 39940853 Free PMC article.
-
A programmable pAgo nuclease with RNA target-cleavage specificity from the mesophilic bacterium Verrucomicrobia.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 10;55(8):1204-1212. doi: 10.3724/abbs.2023110. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37431184 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

