Pseudomonas aeruginosa Bacterioferritin Is Assembled from FtnA and BfrB Subunits with the Relative Proportions Dependent on the Environmental Oxygen Availability
- PMID: 35327558
- PMCID: PMC8945002
- DOI: 10.3390/biom12030366
Pseudomonas aeruginosa Bacterioferritin Is Assembled from FtnA and BfrB Subunits with the Relative Proportions Dependent on the Environmental Oxygen Availability
Abstract
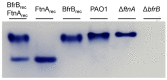
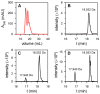
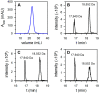
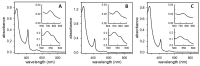
Ferritins are iron storage proteins assembled from 24 subunits into a spherical and hollow structure. The genomes of many bacteria harbor genes encoding two types of ferritin-like proteins, the bacterial ferritins (Ftn) and the bacterioferritins (Bfr), which bind heme. The genome of P. aeruginosa PAO1 (like the genomes of many bacteria) contains genes coding for two different types of ferritin-like molecules, ftnA (PA4235) and bfrB (PA3531). The reasons for requiring the presence of two distinct types of iron storage protein in bacterial cells have remained largely unexplained. Attempts to understand this issue in P. aeruginosa through the recombinant expression of the ftnA and bfrB genes in E. coli host cells, coupled to the biochemical and structural characterization of the recombinant 24-mer FtnA and 24-mer BfrB molecules, have shown that each of the recombinant molecules can form an Fe3+-mineral core. These observations led to the suggestion that 24-mer FtnA and 24-mer BfrB molecules coexist in P. aeruginosa cells where they share iron storage responsibilities. Herein, we demonstrate that P. aeruginosa utilizes a single heterooligomeric 24-mer Bfr assembled from FtnA and BfrB subunits. The relative content of the FtnA and BfrB subunits in Bfr depends on the O2 availability during cell culture, such that Bfr isolated from aerobically cultured P. aeruginosa is assembled from a majority of BfrB subunits. In contrast, when the cells are cultured in O2-limiting conditions, the proportion of FtnA subunits in the isolated Bfr increases significantly and can become the most abundant subunit. Despite the variability in the subunit composition of Bfr, the 24-mer assembly is consistently arranged from FtnA subunit dimers devoid of heme and BfrB subunit dimers each containing a heme molecule.
Keywords: aerobic culture; anaerobic culture; bacterioferritin; ferritin; iron metabolism; iron storage; microaerophilic culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).Biochemistry. 2011 Jun 14;50(23):5236-48. doi: 10.1021/bi2004119. Epub 2011 May 20. Biochemistry. 2011. PMID: 21574546 Free PMC article.
-
Bacterioferritin: Structure, Dynamics, and Protein-Protein Interactions at Play in Iron Storage and Mobilization.Acc Chem Res. 2017 Feb 21;50(2):331-340. doi: 10.1021/acs.accounts.6b00514. Epub 2017 Feb 8. Acc Chem Res. 2017. PMID: 28177216 Free PMC article.
-
Inhibiting the BfrB:Bfd interaction in Pseudomonas aeruginosa causes irreversible iron accumulation in bacterioferritin and iron deficiency in the bacterial cytosol.Metallomics. 2017 Jun 21;9(6):646-659. doi: 10.1039/c7mt00042a. Metallomics. 2017. PMID: 28318006 Free PMC article.
-
Iron storage in bacteria.Adv Microb Physiol. 1998;40:281-351. doi: 10.1016/s0065-2911(08)60134-4. Adv Microb Physiol. 1998. PMID: 9889981 Review.
-
Bacterioferritin nanocage: Structure, biological function, catalytic mechanism, self-assembly and potential applications.Biotechnol Adv. 2022 Dec;61:108057. doi: 10.1016/j.biotechadv.2022.108057. Epub 2022 Nov 1. Biotechnol Adv. 2022. PMID: 36328189 Review.
Cited by
-
Global transcriptional response of Pseudomonas aeruginosa to UVA radiation.Photochem Photobiol Sci. 2024 Nov;23(11):2029-2044. doi: 10.1007/s43630-024-00649-9. Epub 2024 Oct 29. Photochem Photobiol Sci. 2024. PMID: 39470974
-
Unveiling the stochastic nature of human heteropolymer ferritin self-assembly mechanism.Protein Sci. 2024 Aug;33(8):e5104. doi: 10.1002/pro.5104. Protein Sci. 2024. PMID: 38995055 Free PMC article.
-
Genetic and environmental determinants of surface adaptations in Pseudomonas aeruginosa.Microbiology (Reading). 2023 Jun;169(6):001335. doi: 10.1099/mic.0.001335. Microbiology (Reading). 2023. PMID: 37276014 Free PMC article.
-
A Closer Look at the FeS Heme Bonds in Azotobacter vinelandii Bacterioferritin: QM/MM and Local Mode Analysis.J Comput Chem. 2025 Jan 5;46(1):e70012. doi: 10.1002/jcc.70012. J Comput Chem. 2025. PMID: 39749917 Free PMC article.
-
The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.Sci Rep. 2024 Aug 6;14(1):18242. doi: 10.1038/s41598-024-69156-2. Sci Rep. 2024. PMID: 39107474 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

