CNN-XG: A Hybrid Framework for sgRNA On-Target Prediction
- PMID: 35327601
- PMCID: PMC8945678
- DOI: 10.3390/biom12030409
CNN-XG: A Hybrid Framework for sgRNA On-Target Prediction
Abstract
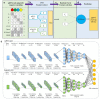
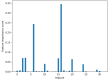
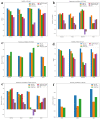
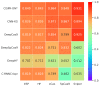
As the third generation gene editing technology, Crispr/Cas9 has a wide range of applications. The success of Crispr depends on the editing of the target gene via a functional complex of sgRNA and Cas9 proteins. Therefore, highly specific and high on-target cleavage efficiency sgRNA can make this process more accurate and efficient. Although there are already many sophisticated machine learning or deep learning models to predict the on-target cleavage efficiency of sgRNA, prediction accuracy remains to be improved. XGBoost is good at classification as the ensemble model could overcome the deficiency of a single classifier to classify, and we would like to improve the prediction efficiency for sgRNA on-target activity by introducing XGBoost into the model. We present a novel machine learning framework which combines a convolutional neural network (CNN) and XGBoost to predict sgRNA on-target knockout efficacy. Our framework, called CNN-XG, is mainly composed of two parts: a feature extractor CNN is used to automatically extract features from sequences and predictor XGBoost is applied to predict features extracted after convolution. Experiments on commonly used datasets show that CNN-XG performed significantly better than other existing frameworks in the predicted classification mode.
Keywords: Crispr/Cas9; XGBoost; deep learning; on-target; sgRNA.
Conflict of interest statement
The authors have no conflict of interest or ethics statement to declare.
Figures
Similar articles
-
sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing.Cell Rep. 2019 Jan 29;26(5):1098-1103.e3. doi: 10.1016/j.celrep.2019.01.024. Cell Rep. 2019. PMID: 30699341 Free PMC article.
-
Leveraging protein language models for cross-variant CRISPR/Cas9 sgRNA activity prediction.Bioinformatics. 2025 Jul 1;41(7):btaf385. doi: 10.1093/bioinformatics/btaf385. Bioinformatics. 2025. PMID: 40600900 Free PMC article.
-
TransCrispr: Transformer Based Hybrid Model for Predicting CRISPR/Cas9 Single Guide RNA Cleavage Efficiency.IEEE/ACM Trans Comput Biol Bioinform. 2023 Mar-Apr;20(2):1518-1528. doi: 10.1109/TCBB.2022.3201631. Epub 2023 Apr 3. IEEE/ACM Trans Comput Biol Bioinform. 2023. PMID: 36006888
-
Points of View on the Tools for Genome/Gene Editing.Int J Mol Sci. 2021 Sep 13;22(18):9872. doi: 10.3390/ijms22189872. Int J Mol Sci. 2021. PMID: 34576035 Free PMC article. Review.
-
Review of CRISPR/Cas9 sgRNA Design Tools.Interdiscip Sci. 2018 Jun;10(2):455-465. doi: 10.1007/s12539-018-0298-z. Epub 2018 Apr 11. Interdiscip Sci. 2018. PMID: 29644494 Review.
Cited by
-
gRNA Design: How Its Evolution Impacted on CRISPR/Cas9 Systems Refinement.Biomolecules. 2023 Nov 24;13(12):1698. doi: 10.3390/biom13121698. Biomolecules. 2023. PMID: 38136570 Free PMC article. Review.
-
Transitioning from wet lab to artificial intelligence: a systematic review of AI predictors in CRISPR.J Transl Med. 2025 Feb 4;23(1):153. doi: 10.1186/s12967-024-06013-w. J Transl Med. 2025. PMID: 39905452 Free PMC article.
-
A systematic mapping study on machine learning techniques for the prediction of CRISPR/Cas9 sgRNA target cleavage.Comput Struct Biotechnol J. 2022 Oct 21;20:5813-5823. doi: 10.1016/j.csbj.2022.10.013. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36382194 Free PMC article. Review.
-
Graph-CRISPR: a gene editing efficiency prediction model based on graph neural network with integrated sequence and secondary structure feature extraction.Brief Bioinform. 2025 Jul 2;26(4):bbaf410. doi: 10.1093/bib/bbaf410. Brief Bioinform. 2025. PMID: 40814228 Free PMC article.
-
DeepMEns: an ensemble model for predicting sgRNA on-target activity based on multiple features.Brief Funct Genomics. 2025 Jan 15;24:elae043. doi: 10.1093/bfgp/elae043. Brief Funct Genomics. 2025. PMID: 39528429 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

