β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation
- PMID: 35327603
- PMCID: PMC8945919
- DOI: 10.3390/biom12030411
β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation
Abstract
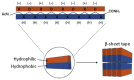
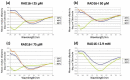
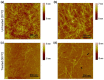
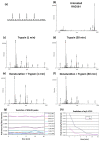
One of the most desirable properties that biomaterials designed for tissue engineering or drug delivery applications should fulfill is biodegradation and resorption without toxicity. Therefore, there is an increasing interest in the development of biomaterials able to be enzymatically degraded once implanted at the injury site or once delivered to the target organ. In this paper, we demonstrate the protease sensitivity of self-assembling amphiphilic peptides, in particular, RAD16-I (AcN-RADARADARADARADA-CONH2), which contains four potential cleavage sites for trypsin. We detected that when subjected to thermal denaturation, the peptide secondary structure suffers a transition from β-sheet to random coil. We also used Matrix-Assisted Laser Desorption/Ionization-Time-Of-Flight (MALDI-TOF) to detect the proteolytic breakdown products of samples subjected to incubation with trypsin as well as atomic force microscopy (AFM) to visualize the effect of the degradation on the nanofiber scaffold. Interestingly, thermally treated samples had a higher extent of degradation than non-denatured samples, suggesting that the transition from β-sheet to random coil leaves the cleavage sites accessible and susceptible to protease degradation. These results indicate that the self-assembling peptide can be reduced to short peptide sequences and, subsequently, degraded to single amino acids, constituting a group of naturally biodegradable materials optimal for their application in tissue engineering and regenerative medicine.
Keywords: RAD16-I; degradation; proteolysis; random coil; scaffold; self-assembling peptides; β-sheet.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Biocompatibility of functionalized designer self-assembling nanofiber scaffolds containing FRM motif for neural stem cells.J Biomed Mater Res A. 2014 May;102(5):1286-93. doi: 10.1002/jbm.a.34804. Epub 2013 Jun 4. J Biomed Mater Res A. 2014. PMID: 23703883
-
[PREPARATION AND BIOCOMPATIBILITY EVALUATION OF A FUNCTIONAL SELF-ASSEMBLING PEPTIDE NANOFIBER HYDROGEL DESIGNED WITH LINKING THE SHORT FUNCTIONAL MOTIF OF BONE MORPHOGENETIC PROTEIN 7].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2016 Apr;30(4):491-8. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2016. PMID: 27411281 Chinese.
-
Design of a RADA16-based self-assembling peptide nanofiber scaffold for biomedical applications.J Biomater Sci Polym Ed. 2019 May-Jun;30(9):713-736. doi: 10.1080/09205063.2019.1605868. Epub 2019 Apr 24. J Biomater Sci Polym Ed. 2019. PMID: 31018781 Review.
-
The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function.Biomaterials. 2005 Jun;26(16):3341-51. doi: 10.1016/j.biomaterials.2004.08.012. Biomaterials. 2005. PMID: 15603830
-
Self-assembling peptides as vectors for local drug delivery and tissue engineering applications.Adv Drug Deliv Rev. 2021 Jul;174:387-405. doi: 10.1016/j.addr.2021.04.024. Epub 2021 May 7. Adv Drug Deliv Rev. 2021. PMID: 33965460 Review.
Cited by
-
MOTS-c-modified functional self-assembly peptide hydrogels enhance the activity of nucleus pulposus-derived mesenchymal stem cells of intervertebral disc degeneration.Mater Today Bio. 2025 May 22;32:101872. doi: 10.1016/j.mtbio.2025.101872. eCollection 2025 Jun. Mater Today Bio. 2025. PMID: 40510834 Free PMC article.
-
Innovative hydrogels in cutaneous wound healing: current status and future perspectives.Front Bioeng Biotechnol. 2025 May 12;13:1454903. doi: 10.3389/fbioe.2025.1454903. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40421113 Free PMC article. Review.
-
Proteolysis of Micellar β-Casein by Trypsin: Secondary Structure Characterization and Kinetic Modeling at Different Enzyme Concentrations.Int J Mol Sci. 2023 Feb 15;24(4):3874. doi: 10.3390/ijms24043874. Int J Mol Sci. 2023. PMID: 36835285 Free PMC article.
-
Increased Stiffness Downregulates Focal Adhesion Kinase Expression in Pancreatic Cancer Cells Cultured in 3D Self-Assembling Peptide Scaffolds.Biomedicines. 2022 Jul 29;10(8):1835. doi: 10.3390/biomedicines10081835. Biomedicines. 2022. PMID: 36009384 Free PMC article.
References
-
- Moffat K.L., Neal R.A., Freed L.E., Guilak F. Engineering Functional Tissues: In Vitro Culture Parameters. In: Lanza R., Langer R., Vacanti J., editors. Principles of Tissue Engineering. 4th ed. Elsevier; Amsterdam, The Netherlands: 2013. pp. 237–259. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

