Dynamical Analysis of a Boolean Network Model of the Oncogene Role of lncRNA ANRIL and lncRNA UFC1 in Non-Small Cell Lung Cancer
- PMID: 35327612
- PMCID: PMC8946683
- DOI: 10.3390/biom12030420
Dynamical Analysis of a Boolean Network Model of the Oncogene Role of lncRNA ANRIL and lncRNA UFC1 in Non-Small Cell Lung Cancer
Abstract
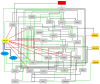
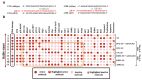
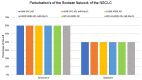
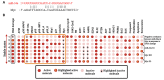
Long non-coding RNA (lncRNA) such as ANRIL and UFC1 have been verified as oncogenic genes in non-small cell lung cancer (NSCLC). It is well known that the tumor suppressor microRNA-34a (miR-34a) is downregulated in NSCLC. Furthermore, miR-34a induces senescence and apoptosis in breast, glioma, cervical cancer including NSCLC by targeting Myc. Recent evidence suggests that these two lncRNAs act as a miR-34a sponge in corresponding cancers. However, the biological functions between these two non-coding RNAs (ncRNAs) have not yet been studied in NSCLC. Therefore, we present a Boolean model to analyze the gene regulation between these two ncRNAs in NSCLC. We compared our model to several experimental studies involving gain- or loss-of-function genes in NSCLC cells and achieved an excellent agreement. Additionally, we predict three positive circuits involving miR-34a/E2F1/ANRIL, miR-34a/E2F1/UFC1, and miR-34a/Myc/ANRIL. Our circuit- perturbation analysis shows that these circuits are important for regulating cell-fate decisions such as senescence and apoptosis. Thus, our Boolean network permits an explicit cell-fate mechanism associated with NSCLC. Therefore, our results support that ANRIL and/or UFC1 is an attractive target for drug development in tumor growth and aggressive proliferation of NSCLC, and that a valuable outcome can be achieved through the miRNA-34a/Myc pathway.
Keywords: ANRIL; Boolean model; Long non-coding RNA; Myc; NSCLC; UFC1; apoptosis; feedback loops; miRNA-34a; senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Long Non-Coding RNA (LncRNA) UFC1/miR-34a Contributes to Proliferation and Migration in Breast Cancer.Med Sci Monit. 2019 Sep 23;25:7149-7157. doi: 10.12659/MSM.917562. Med Sci Monit. 2019. PMID: 31544897 Free PMC article.
-
A Boolean Model of the Proliferative Role of the lncRNA XIST in Non-Small Cell Lung Cancer Cells.Biology (Basel). 2022 Mar 22;11(4):480. doi: 10.3390/biology11040480. Biology (Basel). 2022. PMID: 35453680 Free PMC article.
-
Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a.J Cell Biochem. 2018 Mar;119(3):2708-2718. doi: 10.1002/jcb.26437. Epub 2017 Nov 24. J Cell Biochem. 2018. PMID: 29057547
-
Functions of lncRNA DUXAP8 in non-small cell lung cancer.Mol Biol Rep. 2022 Mar;49(3):2531-2542. doi: 10.1007/s11033-021-07066-6. Epub 2022 Jan 15. Mol Biol Rep. 2022. PMID: 35031926 Review.
-
Comprehensive landscape and future perspective of long noncoding RNAs in non-small cell lung cancer: it takes a village.Mol Ther. 2023 Dec 6;31(12):3389-3413. doi: 10.1016/j.ymthe.2023.09.015. Epub 2023 Sep 21. Mol Ther. 2023. PMID: 37740493 Free PMC article. Review.
Cited by
-
In Silico Pleiotropy Analysis in KEGG Signaling Networks Using a Boolean Network Model.Biomolecules. 2022 Aug 18;12(8):1139. doi: 10.3390/biom12081139. Biomolecules. 2022. PMID: 36009032 Free PMC article.
-
The lncRNA DLX6-AS1/miR-16-5p axis regulates autophagy and apoptosis in non-small cell lung cancer: A Boolean model of cell death.Noncoding RNA Res. 2023 Aug 10;8(4):605-614. doi: 10.1016/j.ncrna.2023.08.003. eCollection 2023 Dec. Noncoding RNA Res. 2023. PMID: 37767112 Free PMC article.
-
Network analysis reveals that the tumor suppressor lncRNA GAS5 acts as a double-edged sword in response to DNA damage in gastric cancer.Sci Rep. 2022 Oct 31;12(1):18312. doi: 10.1038/s41598-022-21492-x. Sci Rep. 2022. PMID: 36316351 Free PMC article.
-
A dynamic Boolean network reveals that the BMI1 and MALAT1 axis is associated with drug resistance by limiting miR-145-5p in non-small cell lung cancer.Noncoding RNA Res. 2023 Oct 19;9(1):185-193. doi: 10.1016/j.ncrna.2023.10.008. eCollection 2024 Mar. Noncoding RNA Res. 2023. PMID: 38125755 Free PMC article.
-
Dynamical modeling of miR-34a, miR-449a, and miR-16 reveals numerous DDR signaling pathways regulating senescence, autophagy, and apoptosis in HeLa cells.Sci Rep. 2022 Mar 22;12(1):4911. doi: 10.1038/s41598-022-08900-y. Sci Rep. 2022. PMID: 35318393 Free PMC article.
References
-
- Nie F., Sun M., Yang J., Xie M., Xu T., Xia R., Liu Y., Liu X., Zhang E., Lu K., et al. Long Noncoding RNA ANRIL Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol. Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. - DOI - PubMed
-
- Zang X., Gu J., Zhang J., Shi H., Hou S., Xu X., Chen Y., Zhang Y., Mao F., Qian H., et al. Exosome-Transmitted LncRNA UFC1 Promotes Non-Small-Cell Lung Cancer Progression by EZH2-Mediated Epigenetic Silencing of PTEN Expression. Cell Death Dis. 2020;11:215. doi: 10.1038/s41419-020-2409-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

