NAMPT Inhibitor and P73 Activator Represses P53 R175H Mutated HNSCC Cell Proliferation in a Synergistic Manner
- PMID: 35327630
- PMCID: PMC8946684
- DOI: 10.3390/biom12030438
NAMPT Inhibitor and P73 Activator Represses P53 R175H Mutated HNSCC Cell Proliferation in a Synergistic Manner
Abstract
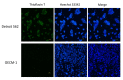
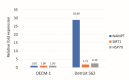
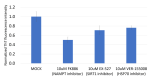
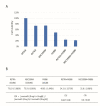
The p53 family has the following three members: p53, p63 and p73. p53 is a tumor suppressor gene that frequently exhibits mutation in head and neck cancer. Most p53 mutants are loss-of-function (LoF) mutants, but some acquire some oncogenic function, such as gain of function (GoF). It is known that the aggregation of mutant p53 can induce p53 GoF. The p73 activators RETRA and NSC59984 have an anti-cancer effect in p53 mutation cells, but we found that p73 activators were not effective in all head and neck squamous cell carcinoma (HNSCC) cell lines, with different p53 mutants. A comparison of the gene expression profiles of several regulator(s) in mutant HNSCC cells with or without aggregation of p53 revealed that nicotinamide phosphoribosyltransferase (NAMPT) is a key regulator of mutant p53 aggregation. An NAMPT inhibitor, to reduce abnormal aggregation of mutant p53, used in combination with a p73 activator, was able to effectively repress growth in HNSCC cells with p53 GoF mutants. This study, therefore, suggests a potential combination therapy approach for HNSCC with a p53 GoF mutation.
Keywords: NAMPT; aggregation; hand and neck; p53; p73.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
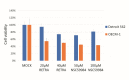
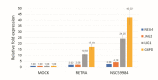
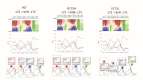
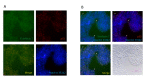
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

