Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins
- PMID: 35327656
- PMCID: PMC8946245
- DOI: 10.3390/biom12030464
Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins
Abstract
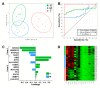
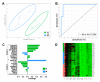
Relapse after surgery for oral squamous cell carcinoma (OSCC) contributes significantly to morbidity, mortality and poor outcomes. The current histopathological diagnostic techniques are insufficiently sensitive for the detection of oral cancer and minimal residual disease in surgical margins. We used whole-transcriptome gene expression and small noncoding RNA profiles from tumour, close margin and distant margin biopsies from 18 patients undergoing surgical resection for OSCC. By applying multivariate regression algorithms (sPLS-DA) suitable for higher dimension data, we objectively identified biomarker signatures for tumour and marginal tissue zones. We were able to define molecular signatures that discriminated tumours from the marginal zones and between the close and distant margins. These signatures included genes not previously associated with OSCC, such as MAMDC2, SYNPO2 and ARMH4. For discrimination of the normal and tumour sampling zones, we were able to derive an effective gene-based classifying model for molecular abnormality based on a panel of eight genes (MMP1, MMP12, MYO1B, TNFRSF12A, WDR66, LAMC2, SLC16A1 and PLAU). We demonstrated the classification performance of these gene signatures in an independent validation dataset of OSCC tumour and marginal gene expression profiles. These biomarker signatures may contribute to the earlier detection of tumour cells and complement existing surgical and histopathological techniques used to determine clear surgical margins.
Keywords: gene biomarker signature; gene expression profiling; multivariate statistics; oral squamous cell carcinoma; surgical margins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gatta G., Botta L., Sánchez M.J., Anderson L.A., Pierannunzio D., Licitra L., on behalf of the EUROCARE Working Group Prognoses and Improvement for Head and Neck Cancers Diagnosed in Europe in Early 2000s: The EUROCARE-5 Population-Based Study. Eur. J. Cancer. 2015;51:2130–2143. doi: 10.1016/j.ejca.2015.07.043. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

