A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations
- PMID: 35327657
- PMCID: PMC8946084
- DOI: 10.3390/biom12030465
A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations
Abstract
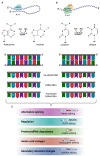
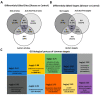
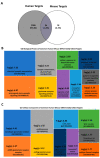
RNA editing contributes to transcriptome diversification through RNA modifications in relation to genome-encoded information (RNA-DNA differences, RDDs). The deamination of Adenosine (A) to Inosine (I) or Cytidine (C) to Uridine (U) is the most common type of mammalian RNA editing. It occurs as a nuclear co- and/or post-transcriptional event catalyzed by ADARs (Adenosine deaminases acting on RNA) and APOBECs (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like genes). RNA editing may modify the structure, stability, and processing of a transcript. This review focuses on RNA editing in psychiatric, neurological, neurodegenerative (NDs), and autoimmune brain disorders in humans and rodent models. We discuss targeted studies that focus on RNA editing in specific neuron-enriched transcripts with well-established functions in neuronal activity, and transcriptome-wide studies, enabled by recent technological advances. We provide comparative editome analyses between human disease and corresponding animal models. Data suggest RNA editing to be an emerging mechanism in disease development, displaying common and disease-specific patterns. Commonly edited RNAs represent potential disease-associated targets for therapeutic and diagnostic values. Currently available data are primarily descriptive, calling for additional research to expand global editing profiles and to provide disease mechanistic insights. The potential use of RNA editing events as disease biomarkers and available tools for RNA editing identification, classification, ranking, and functional characterization that are being developed will enable comprehensive analyses for a better understanding of disease(s) pathogenesis and potential cures.
Keywords: RNA editing; brain disorders; neurodegenerative diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A novel mechanism for A-to-I RNA-edited CYP1A1 in promoting cancer progression in NSCLC.Cell Mol Biol Lett. 2025 Apr 2;30(1):40. doi: 10.1186/s11658-025-00718-6. Cell Mol Biol Lett. 2025. PMID: 40175891 Free PMC article.
-
Structural analysis of human ADAR2-RNA complexes by X-ray crystallography.Methods Enzymol. 2025;710:19-53. doi: 10.1016/bs.mie.2024.11.023. Epub 2024 Dec 4. Methods Enzymol. 2025. PMID: 39870445 Free PMC article.
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
-
Inflammation primes the murine kidney for recovery by activating AZIN1 adenosine-to-inosine editing.J Clin Invest. 2024 Sep 3;134(17):e180117. doi: 10.1172/JCI180117. J Clin Invest. 2024. PMID: 38954486 Free PMC article.
Cited by
-
Comprehensive characterization of the RNA editing landscape in the human aging brains with Alzheimer's disease.Alzheimers Dement. 2025 Jul;21(7):e70452. doi: 10.1002/alz.70452. Alzheimers Dement. 2025. PMID: 40631452 Free PMC article.
-
The coevolution between APOBEC3 and retrotransposons in primates.Mob DNA. 2022 Nov 29;13(1):27. doi: 10.1186/s13100-022-00283-1. Mob DNA. 2022. PMID: 36443831 Free PMC article. Review.
-
The interplay between epitranscriptomic RNA modifications and neurodegenerative disorders: Mechanistic insights and potential therapeutic strategies.Ibrain. 2024 Nov 11;10(4):395-426. doi: 10.1002/ibra.12183. eCollection 2024 Winter. Ibrain. 2024. PMID: 39691424 Free PMC article. Review.
-
In silico detection of dysregulated genes and molecular pathways in Alzheimer's disease as basis for food restoring approach.PeerJ. 2025 Apr 7;13:e19100. doi: 10.7717/peerj.19100. eCollection 2025. PeerJ. 2025. PMID: 40212376 Free PMC article.
-
RNA epitranscriptomics dysregulation: A major determinant for significantly increased risk of ASD pathogenesis.Front Neurosci. 2023 Feb 16;17:1101422. doi: 10.3389/fnins.2023.1101422. eCollection 2023. Front Neurosci. 2023. PMID: 36875672 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

