Identification and Characterization of Circular RNAs in Mammary Tissue from Holstein Cows at Early Lactation and Non-Lactation
- PMID: 35327670
- PMCID: PMC8946036
- DOI: 10.3390/biom12030478
Identification and Characterization of Circular RNAs in Mammary Tissue from Holstein Cows at Early Lactation and Non-Lactation
Abstract
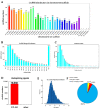
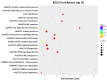
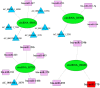
In this study, circular RNAs (circRNAs) from Holstein cow mammary tissues were identified and compared between early lactation and non-lactation. After analysis, 10,684 circRNAs were identified, ranging from 48 to 99,406 bp, and the average size was 882 bp. The circRNAs were mainly distributed on chromosomes 1 to 11, and 89.89% of the circRNAs belonged to sense-overlapping circRNA. The exons contained with circRNAs ranged from 1 to 47 and were concentrated from 1 to 5. Compared with the non-lactating cows, 87 circRNAs were significantly differentially expressed in the peak lactation cows. There were 68 upregulated circRNAs and 19 downregulated circRNAs. Enrichment analysis of circRNAs showed that GO analysis mainly focused on immune response, triglyceride transport, T cell receptor signaling pathway, etc. Pathway analysis mainly focused on cytokine-cytokine receptor interaction, T helper 17 cell differentiation, fatty acid biosynthesis, the JAK-STAT signaling pathway, etc. Specific primers were designed for two proximal ends of the circRNA junction sites to allow for PCR validation of four randomly selected circRNAs and carry out circRNA-miRNA interaction research. This study revealed the expression profile and characteristics of circRNAs in mammary tissue from Holstein cows at early lactation and non-lactation, thus providing rich information for the study of circRNA functions and mechanisms, as well as potential candidate miRNA genes for studying lactation in Holstein cows.
Keywords: Holstein cows; RNA sequencing; circular RNA; mammary tissue.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Developmental Regulation of circRNAs in Normal and Diseased Mammary Gland: A Focus on circRNA-miRNA Networks.J Mammary Gland Biol Neoplasia. 2025 May 2;30(1):8. doi: 10.1007/s10911-025-09580-w. J Mammary Gland Biol Neoplasia. 2025. PMID: 40314719 Free PMC article. Review.
-
Excavation and characterization of key circRNAs for milk fat percentage in Holstein cattle.J Anim Sci. 2023 Jan 3;101:skad157. doi: 10.1093/jas/skad157. J Anim Sci. 2023. PMID: 37209411 Free PMC article.
-
High-throughput analysis of CircRNA in cows with naturally infected Staphylococcus aureus mammary gland.Anim Biotechnol. 2023 Dec;34(9):4236-4246. doi: 10.1080/10495398.2022.2140056. Epub 2022 Dec 28. Anim Biotechnol. 2023. PMID: 36576137
-
Identification and characterization of long non-coding RNAs in mammary gland tissues of Chinese Holstein cows.J Anim Sci. 2024 Jan 3;102:skae128. doi: 10.1093/jas/skae128. J Anim Sci. 2024. PMID: 38715467 Free PMC article.
-
Transcriptomics reveals the regulatory mechanisms of circRNA in the muscle tissue of cows with ketosis postpartum.Genomics. 2025 Mar;117(2):111008. doi: 10.1016/j.ygeno.2025.111008. Epub 2025 Jan 22. Genomics. 2025. PMID: 39855483 Review.
Cited by
-
Non-coding transcriptomic profiles in the sheep mammary gland during different lactation periods.Front Vet Sci. 2022 Nov 8;9:983562. doi: 10.3389/fvets.2022.983562. eCollection 2022. Front Vet Sci. 2022. PMID: 36425117 Free PMC article.
-
Developmental Regulation of circRNAs in Normal and Diseased Mammary Gland: A Focus on circRNA-miRNA Networks.J Mammary Gland Biol Neoplasia. 2025 May 2;30(1):8. doi: 10.1007/s10911-025-09580-w. J Mammary Gland Biol Neoplasia. 2025. PMID: 40314719 Free PMC article. Review.
-
Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period.Animals (Basel). 2025 Jan 3;15(1):89. doi: 10.3390/ani15010089. Animals (Basel). 2025. PMID: 39795032 Free PMC article.
-
Tissue-Specific Expression of Circ_015343 and Its Inhibitory Effect on Mammary Epithelial Cells in Sheep.Front Vet Sci. 2022 Jun 28;9:919162. doi: 10.3389/fvets.2022.919162. eCollection 2022. Front Vet Sci. 2022. PMID: 35836501 Free PMC article.
-
Understanding Circular RNAs in Health, Welfare, and Productive Traits of Cattle, Goats, and Sheep.Animals (Basel). 2024 Feb 27;14(5):733. doi: 10.3390/ani14050733. Animals (Basel). 2024. PMID: 38473119 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

