Alternative Splicing and Isoforms: From Mechanisms to Diseases
- PMID: 35327956
- PMCID: PMC8951537
- DOI: 10.3390/genes13030401
Alternative Splicing and Isoforms: From Mechanisms to Diseases
Abstract
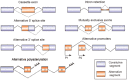
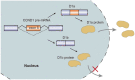
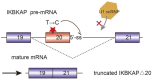
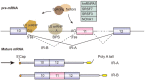
Alternative splicing of pre-mRNA is a key mechanism for increasing the complexity of proteins in humans, causing a diversity of expression of transcriptomes and proteomes in a tissue-specific manner. Alternative splicing is regulated by a variety of splicing factors. However, the changes and errors of splicing regulation caused by splicing factors are strongly related to many diseases, something which represents one of this study's main interests. Further understanding of alternative splicing regulation mediated by cellular factors is also a prospective choice to develop specific drugs for targeting the dynamic RNA splicing process. In this review, we firstly concluded the basic principle of alternative splicing. Afterwards, we showed how splicing isoforms affect physiological activities through specific disease examples. Finally, the available treatment methods relative to adjusting splicing activities have been summarized.
Keywords: alternative splicing; diseases; drugs; splicing factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

