Conserved Molecular Signatures in the Spike, Nucleocapsid, and Polymerase Proteins Specific for the Genus Betacoronavirus and Its Different Subgenera
- PMID: 35327976
- PMCID: PMC8949385
- DOI: 10.3390/genes13030423
Conserved Molecular Signatures in the Spike, Nucleocapsid, and Polymerase Proteins Specific for the Genus Betacoronavirus and Its Different Subgenera
Abstract
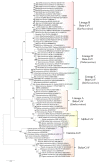
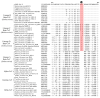
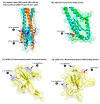
The genus Betacoronavirus, consisting of four main subgenera (Embecovirus, Merbecovirus, Nobecovirus, and Sarbecovirus), encompasses all clinically significant coronaviruses (CoVs), including SARS, MERS, and the SARS-CoV-2 virus responsible for current COVID-19 pandemic. Very few molecular characteristics are known that are specific for the genus Betacoronavirus or its different subgenera. In this study, our analyses of the sequences of four essential proteins of CoVs, viz., spike, nucleocapsid, envelope, and RNA-dependent RNA polymerase (RdRp), identified ten novel molecular signatures consisting of conserved signature indels (CSIs) in these proteins which are specific for the genus Betacoronavirus or its subgenera. Of these CSIs, two 14-aa-conserved deletions found within the heptad repeat motifs 1 and 2 of the spike protein are specific for all betacoronaviruses, except for their shared presence in the highly infectious avian coronavirus. Six additional CSIs present in the nucleocapsid protein and one CSI in the RdRp protein are distinctive characteristics of either the Merbecovirus, Nobecovirus, or Sarbecovirus subgenera. In addition, a 4-aa insert is present in the spike protein, which is uniquely shared by all viruses from the subgenera Merbecovirus, Nobecovirus, and Sarbecovirus, but absent in Embecovirus and all other genera of CoVs. This molecular signature provides evidence that viruses from the three subgenera sharing this CSI are more closely related to each other, and they evolved after the divergence of embecoviruses and other CoVs. As all CSIs specific for different groups of CoVs are flanked by conserved regions, their sequences provide novel means for identifying the above groups of CoVs and for developing novel diagnostic tests. Furthermore, our analyses of the structures of the spike and nucleocapsid proteins show that all identified CSIs are localized in the surface-exposed loops of these protein. It is postulated that these surface loops, through their interactions with other cellular proteins/ligands, play important roles in the biology/pathology of these viruses.
Keywords: Betacoronavirus and its subgenera Sarbecovirus; Merbecovirus and Nobecovirus; conserved signature indels (CSIs); evolution of betacoronaviruses; heptad repeat motifs 1 and 2; molecular markers; nucleocapsid and RNA-dependent RNA polymerase (RdRp) proteins; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Conserved molecular signatures in the spike protein provide evidence indicating the origin of SARS-CoV-2 and a Pangolin-CoV (MP789) by recombination(s) between specific lineages of Sarbecoviruses.PeerJ. 2021 Nov 12;9:e12434. doi: 10.7717/peerj.12434. eCollection 2021. PeerJ. 2021. PMID: 35028194 Free PMC article.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
Recombination and Positive Selection Differentially Shaped the Diversity of Betacoronavirus Subgenera.Viruses. 2020 Nov 16;12(11):1313. doi: 10.3390/v12111313. Viruses. 2020. PMID: 33207802 Free PMC article.
-
Novel Molecular Synapomorphies Demarcate Different Main Groups/Subgroups of Plasmodium and Piroplasmida Species Clarifying Their Evolutionary Relationships.Genes (Basel). 2019 Jun 28;10(7):490. doi: 10.3390/genes10070490. Genes (Basel). 2019. PMID: 31261747 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Recombination analysis on the receptor switching event of MERS-CoV and its close relatives: implications for the emergence of MERS-CoV.Virol J. 2024 Apr 10;21(1):84. doi: 10.1186/s12985-024-02358-2. Virol J. 2024. PMID: 38600521 Free PMC article.
-
ACE2-using merbecoviruses: Further evidence of convergent evolution of ACE2 recognition by NeoCoV and other MERS-CoV related viruses.Cell Insight. 2024 Jan 30;3(1):100145. doi: 10.1016/j.cellin.2023.100145. eCollection 2024 Feb. Cell Insight. 2024. PMID: 38476250 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

