Comparative Transcriptomic Analysis of Hu Sheep Pituitary Gland Prolificacy at the Follicular and Luteal Phases
- PMID: 35327994
- PMCID: PMC8949571
- DOI: 10.3390/genes13030440
Comparative Transcriptomic Analysis of Hu Sheep Pituitary Gland Prolificacy at the Follicular and Luteal Phases
Abstract
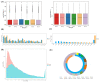
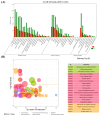
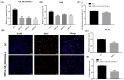
The pituitary gland directly regulates the reproduction of domestic animals. Research has increasingly focused on the potential regulatory mechanism of non-coding RNA in pituitary development. Little is known about the differential expression pattern of lncRNAs in Hu sheep, a famous sheep breed with high fecundity, and its role in the pituitary gland between the follicular phase and luteal phase. Herein, to identify the transcriptomic differences of the sheep pituitary gland during the estrus cycle, RNA sequencing (RNA-Seq) was performed. The results showed that 3529 lncRNAs and 16,651 mRNAs were identified in the pituitary gland. Among of them, 144 differentially expressed (DE) lncRNA transcripts and 557 DE mRNA transcripts were screened in the follicular and luteal phases. Moreover, GO and KEGG analyses demonstrated that 39 downregulated and 22 upregulated genes interacted with pituitary functions and reproduction. Lastly, the interaction of the candidate lncRNA XR_001039544.4 and its targeted gene LHB were validated in sheep pituitary cells in vitro. LncRNA XR_001039544.4 and LHB showed high expression levels in the luteal phase in Hu sheep. LncRNA XR_001039544.4 is mainly located in the cytoplasm, as determined by FISH analysis, indicating that XR_001039544.4 might act as competing endogenous RNAs for miRNAs to regulate LHB. LncRNA XR_001039544.4 knockdown significantly inhibited LH secretion and cell proliferation. LncRNA XR_001039544.4 may regulate the secretion of LH in the luteal-phase pituitary gland via affecting cell proliferation. Taken together, these findings provided genome-wide lncRNA- and mRNA-expression profiles for the sheep pituitary gland between the follicular and luteal phases, thereby contributing to the elucidation of the molecular mechanisms of pituitary function.
Keywords: LHB; fecundity; follicular phase; lncRNA; luteal phase; pituitary gland.
Conflict of interest statement
The authors have declared that they have no conflict of interest.
Figures






Similar articles
-
Pineal gland transcriptomic profiling reveals the differential regulation of lncRNA and mRNA related to prolificacy in STH sheep with two FecB genotypes.BMC Genom Data. 2021 Feb 18;22(1):9. doi: 10.1186/s12863-020-00957-w. BMC Genom Data. 2021. PMID: 33602139 Free PMC article.
-
Expression profiles of oviductal mRNAs and lncRNAs in the follicular phase and luteal phase of sheep (Ovis aries) with 2 fecundity gene (FecB) genotypes.G3 (Bethesda). 2023 Dec 29;14(1):jkad270. doi: 10.1093/g3journal/jkad270. G3 (Bethesda). 2023. PMID: 38051961 Free PMC article.
-
Pituitary Transcriptomic Study Reveals the Differential Regulation of lncRNAs and mRNAs Related to Prolificacy in Different FecB Genotyping Sheep.Genes (Basel). 2019 Feb 18;10(2):157. doi: 10.3390/genes10020157. Genes (Basel). 2019. PMID: 30781725 Free PMC article.
-
Key mRNAs and lncRNAs of pituitary that affect the reproduction of FecB + + small tail han sheep.BMC Genomics. 2024 Apr 22;25(1):392. doi: 10.1186/s12864-024-10191-8. BMC Genomics. 2024. PMID: 38649819 Free PMC article.
-
Genome-wide differential expression profiling of mRNAs and lncRNAs associated with prolificacy in Hu sheep.Biosci Rep. 2018 Apr 27;38(2):BSR20171350. doi: 10.1042/BSR20171350. Print 2018 Apr 27. Biosci Rep. 2018. PMID: 29439142 Free PMC article.
Cited by
-
Transcriptome analysis reveals pituitary lncRNA, circRNA and mRNA affecting fertility in high- and low-yielding goats.Front Genet. 2023 Dec 12;14:1303031. doi: 10.3389/fgene.2023.1303031. eCollection 2023. Front Genet. 2023. PMID: 38152654 Free PMC article.
-
Chromatin accessibility and transcriptomic profiles of sheep pituitary function associated with fecundity.BMC Genomics. 2025 May 20;26(1):508. doi: 10.1186/s12864-025-11621-x. BMC Genomics. 2025. PMID: 40394458 Free PMC article.
-
EZH2 Gene Knockdown Inhibits Sheep Pituitary Cell Proliferation via Downregulating the AKT/ERK Signaling Pathway.Int J Mol Sci. 2023 Jun 26;24(13):10656. doi: 10.3390/ijms241310656. Int J Mol Sci. 2023. PMID: 37445833 Free PMC article.
-
Metagenomics-Metabolomics Exploration of Three-Way-Crossbreeding Effects on Rumen to Provide Basis for Crossbreeding Improvement of Sheep Microbiome and Metabolome of Sheep.Animals (Basel). 2024 Aug 3;14(15):2256. doi: 10.3390/ani14152256. Animals (Basel). 2024. PMID: 39123781 Free PMC article.
-
Integration analysis of pituitary proteome and transcriptome reveals fertility-related biomarkers in FecB mutant Small Tail Han sheep.Front Endocrinol (Lausanne). 2024 Jul 23;15:1417530. doi: 10.3389/fendo.2024.1417530. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39109077 Free PMC article.
References
-
- Wilson T., Wu X.Y., Juengel J.L., Ross I.K., Lumsden J.M., Lord E.A., Dodds K.G., Walling G.A., McEwan J.C., O’Connell A.R., et al. Highly prolific booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. - DOI - PubMed
-
- Mulsant P., Lecerf F., Fabre S., Schibler L., Monget P., Lanneluc I., Pisselet C., Riquet J., Monniaux D., Callebaut I., et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. USA. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

