Transcriptome Profiling Reveals Role of MicroRNAs and Their Targeted Genes during Adventitious Root Formation in Dark-Pretreated Micro-Shoot Cuttings of Tetraploid Robinia pseudoacacia L
- PMID: 35327995
- PMCID: PMC8950900
- DOI: 10.3390/genes13030441
Transcriptome Profiling Reveals Role of MicroRNAs and Their Targeted Genes during Adventitious Root Formation in Dark-Pretreated Micro-Shoot Cuttings of Tetraploid Robinia pseudoacacia L
Abstract
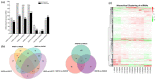
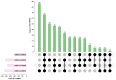
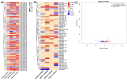
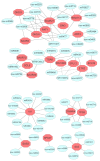
Tetraploid Robinia pseudoacacia L. is a difficult-to-root species, and is vegetatively propagated through stem cuttings. Limited information is available regarding the adventitious root (AR) formation of dark-pretreated micro-shoot cuttings. Moreover, the role of specific miRNAs and their targeted genes during dark-pretreated AR formation under in vitro conditions has never been revealed. The dark pretreatment has successfully promoted and stimulated adventitious rooting signaling-related genes in tissue-cultured stem cuttings with the application of auxin (0.2 mg L-1 IBA). Histological analysis was performed for AR formation at 0, 12, 36, 48, and 72 h after excision (HAE) of the cuttings. The first histological events were observed at 36 HAE in the dark-pretreated cuttings; however, no cellular activities were observed in the control cuttings. In addition, the present study aimed to uncover the role of differentially expressed (DE) microRNAs (miRNAs) and their targeted genes during adventitious root formation using the lower portion (1-1.5 cm) of tetraploid R. pseudoacacia L. micro-shoot cuttings. The samples were analyzed using Illumina high-throughput sequencing technology for the identification of miRNAs at the mentioned time points. Seven DE miRNA libraries were constructed and sequenced. The DE number of 81, 162, 153, 154, 41, 9, and 77 miRNAs were upregulated, whereas 67, 98, 84, 116, 19, 16, and 93 miRNAs were downregulated in the following comparisons of the libraries: 0-vs-12, 0-vs-36, 0-vs-48, 0-vs-72, 12-vs-36, 36-vs-48, and 48-vs-72, respectively. Furthermore, we depicted an association between ten miRNAs (novel-m0778-3p, miR6135e.2-5p, miR477-3p, miR4416c-5p, miR946d, miR398b, miR389a-3p, novel m0068-5p, novel-m0650-3p, and novel-m0560-3p) and important target genes (auxin response factor-3, gretchen hagen-9, scarecrow-like-1, squamosa promoter-binding protein-like-12, small auxin upregulated RNA-70, binding protein-9, vacuolar invertase-1, starch synthase-3, sucrose synthase-3, probable starch synthase-3, cell wall invertase-4, and trehalose phosphatase synthase-5), all of which play a role in plant hormone signaling and starch and sucrose metabolism pathways. The quantitative polymerase chain reaction (qRT-PCR) was used to validate the relative expression of these miRNAs and their targeted genes. These results provide novel insights and a foundation for further studies to elucidate the molecular factors and processes controlling AR formation in woody plants.
Keywords: RNA-seq; adventitious rooting; dark pretreatment; miRNA-seq; tetraploid Robinia pseudoacacia L..
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Temporal profiling of physiological, histological, and transcriptomic dissection during auxin-induced adventitious root formation in tetraploid Robinia pseudoacacia micro-cuttings.Planta. 2024 Feb 8;259(3):66. doi: 10.1007/s00425-024-04341-1. Planta. 2024. PMID: 38332379
-
De novo sequencing and comparative transcriptome analysis of adventitious root development induced by exogenous indole-3-butyric acid in cuttings of tetraploid black locust.BMC Genomics. 2017 Feb 16;18(1):179. doi: 10.1186/s12864-017-3554-4. BMC Genomics. 2017. PMID: 28209181 Free PMC article.
-
Transcriptomic profiling and discovery of key genes involved in adventitious root formation from green cuttings of highbush blueberry (Vaccinium corymbosum L.).BMC Plant Biol. 2020 Apr 25;20(1):182. doi: 10.1186/s12870-020-02398-0. BMC Plant Biol. 2020. PMID: 32334538 Free PMC article.
-
Auxin-Induced Adventitious Root Formation in Nodal Cuttings of Camellia sinensis.Int J Mol Sci. 2019 Sep 27;20(19):4817. doi: 10.3390/ijms20194817. Int J Mol Sci. 2019. PMID: 31569758 Free PMC article. Review.
-
Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings.Front Plant Sci. 2016 Mar 31;7:381. doi: 10.3389/fpls.2016.00381. eCollection 2016. Front Plant Sci. 2016. PMID: 27064322 Free PMC article. Review.
Cited by
-
Exogenous hormones supplementation improve adventitious root formation in woody plants.Front Bioeng Biotechnol. 2022 Sep 13;10:1009531. doi: 10.3389/fbioe.2022.1009531. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36177185 Free PMC article. No abstract available.
-
High-temperature stress in crops: male sterility, yield loss and potential remedy approaches.Plant Biotechnol J. 2023 Apr;21(4):680-697. doi: 10.1111/pbi.13946. Epub 2022 Nov 4. Plant Biotechnol J. 2023. PMID: 36221230 Free PMC article. Review.
-
The Targeted Metabolomic Signatures of Phytohormones in Leaves of Mulberry (Morus alba L.) Are Crucial for Regrowth and Specifically Modulated by the Differential Stubble Lengths.Plants (Basel). 2025 Apr 5;14(7):1126. doi: 10.3390/plants14071126. Plants (Basel). 2025. PMID: 40219194 Free PMC article.
-
Transcriptomic and Phenotypic Analyses Reveal the Molecular Mechanism of Dwarfing in Tetraploid Robinia pseudoacacia L.Int J Mol Sci. 2024 Jan 21;25(2):1312. doi: 10.3390/ijms25021312. Int J Mol Sci. 2024. PMID: 38279314 Free PMC article.
-
The mechanism of PGR-free direct organogenesis in Lycium ruthenicum leaf explants revealed by RNA-Seq and phytohormone metabolome co-analysis.BMC Plant Biol. 2025 Jul 2;25(1):825. doi: 10.1186/s12870-025-06835-w. BMC Plant Biol. 2025. PMID: 40604462 Free PMC article.
References
-
- Zhang G., Li Y., Li F., Xu Z., Sun Y. Effects of root age on biomass and leaf nutrition in tetraploid Robinia pseudoacacia. J. Beijing For. Univ. 2009;31:37–41.
-
- Zhang S., Zhao Z., Zhang L., Zhou Q. Comparative proteomic analysis of tetraploid black locust (Robinia pseudoacacia L.) cuttings in different phases of adventitious root development. Trees. 2015;29:367–384. doi: 10.1007/s00468-014-1116-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

