Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China
- PMID: 35327998
- PMCID: PMC8954247
- DOI: 10.3390/genes13030444
Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China
Abstract
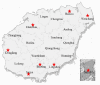
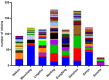
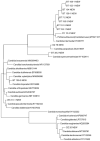
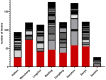
Yeasts play important roles in both the environment and in human welfare. While some environmental yeasts positively contribute to nutrient cycling and food production, a significant number of yeast species are opportunistic human pathogens, including several that are tolerant/resistant to commonly used antifungal drugs. At present, most of our understanding of environmental yeasts has come from a few terrestrial environments in selected geographic regions. Relatively little is known about yeast diversity in tropical environments and their potential impacts on human health. Here, we characterize culturable yeasts in 968 environmental samples from eight regions in tropical China. Among the 516 soil, 273 freshwater, and 179 seawater samples, 71.5%, 85.7%, and 43.6% contained yeasts, respectively. A total of 984 yeast isolates were analyzed for their DNA barcode sequences and their susceptibilities to fluconazole. DNA sequence comparisons revealed that the 984 yeast isolates likely belonged to 144 species, including 106 known species and 38 putative novel species. About 38% of the 984 isolates belonged to known human pathogens and the most common species was Candida tropicalis, accounting for 21% (207/984) of all isolates. Further analyses based on multi-locus sequence typing revealed that some of these environmental C. tropicalis shared identical genotypes with clinical isolates previously reported from tropical China and elsewhere. Importantly, 374 of the 984 (38%) yeast isolates showed intermediate susceptibility or resistance to fluconazole. Our results suggest that these environmental yeasts could have significant negative impacts on human health.
Keywords: Candida; Candida tropicalis; Rhodotorula; environment yeast; fluconazole-resistance.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Genetic relatedness among azole-resistant Candida tropicalis clinical strains in Taiwan from 2014 to 2018.Int J Antimicrob Agents. 2022 Jun;59(6):106592. doi: 10.1016/j.ijantimicag.2022.106592. Epub 2022 Apr 20. Int J Antimicrob Agents. 2022. PMID: 35460852
-
Multilocus sequence analyses reveal extensive diversity and multiple origins of fluconazole resistance in Candida tropicalis from tropical China.Sci Rep. 2017 Feb 10;7:42537. doi: 10.1038/srep42537. Sci Rep. 2017. PMID: 28186162 Free PMC article.
-
In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study.J Clin Microbiol. 2012 Dec;50(12):3952-9. doi: 10.1128/JCM.01130-12. Epub 2012 Oct 3. J Clin Microbiol. 2012. PMID: 23035204 Free PMC article.
-
Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment.Dan Med J. 2016 Oct;63(10):B5288. Dan Med J. 2016. PMID: 27697142 Review.
-
Candida and candidaemia. Susceptibility and epidemiology.Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
Cited by
-
An agricultural triazole induces genomic instability and haploid cell formation in the human fungal pathogen Candida tropicalis.PLoS Biol. 2025 Apr 1;23(4):e3003062. doi: 10.1371/journal.pbio.3003062. eCollection 2025 Apr. PLoS Biol. 2025. PMID: 40168394 Free PMC article.
-
Water Quality, Heavy Metals, and Antifungal Susceptibility to Fluconazole of Yeasts from Water Systems.Int J Environ Res Public Health. 2023 Feb 15;20(4):3428. doi: 10.3390/ijerph20043428. Int J Environ Res Public Health. 2023. PMID: 36834128 Free PMC article.
-
Assessing global fungal threats to humans.mLife. 2022 Sep 22;1(3):223-240. doi: 10.1002/mlf2.12036. eCollection 2022 Sep. mLife. 2022. PMID: 38818220 Free PMC article. Review.
-
A case report of childhood onychomycosis caused by the rare yeast Kodamaea ohmeri.Med Mycol Case Rep. 2025 Jan 20;47:100695. doi: 10.1016/j.mmcr.2025.100695. eCollection 2025 Mar. Med Mycol Case Rep. 2025. PMID: 39926298 Free PMC article.
-
The emerging threat antifungal-resistant Candida tropicalis in humans, animals, and environment.Front Fungal Biol. 2022 Aug 15;3:957021. doi: 10.3389/ffunb.2022.957021. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746212 Free PMC article. Review.
References
-
- Fell J.W. Collection and identification of marine yeasts. Methods Microbiol. 2001;30:347–356. doi: 10.1016/S0580-9517(01)30052-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

