Characterization of a Read-through Fusion Transcript, BCL2L2-PABPN1, Involved in Porcine Adipogenesis
- PMID: 35327999
- PMCID: PMC8955228
- DOI: 10.3390/genes13030445
Characterization of a Read-through Fusion Transcript, BCL2L2-PABPN1, Involved in Porcine Adipogenesis
Abstract
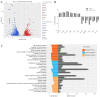
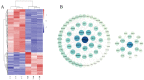
cis-Splicing of adjacent genes (cis-SAGe) has been involved in multiple physiological and pathological processes in humans. However, to the best of our knowledge, there is no report of cis-SAGe in adipogenic regulation. In this study, a cis-SAGe product, BCL2L2-PABPN1 (BP), was characterized in fat tissue of pigs with RT-PCR and RACE method. BP is an in-frame fusion product composed of 333 aa and all the functional domains of both parents. BP is highly conserved among species and rich in splicing variants. BP was found to promote proliferation and inhibit differentiation of primary porcine preadipocytes. A total of 3074/44 differentially expressed mRNAs (DEmRs)/known miRNAs (DEmiRs) were identified in porcine preadipocytes overexpressing BP through RNA-Seq analysis. Both DEmRs and target genes of DEmiRs were involved in various fat-related pathways with MAPK and PI3K-Akt being the top enriched. PPP2CB, EGFR, Wnt5A and EHHADH were hub genes among the fat-related pathways identified. Moreover, ssc-miR-339-3p was found to be critical for BP regulating adipogenesis through integrated analysis of mRNA and miRNA data. The results highlight the role of cis-SAGe in adipogenesis and contribute to further revealing the mechanisms underlying fat deposition, which will be conductive to human obesity control.
Keywords: BCL2L2-PABPN1; RNA-Seq; adipogenesis; chimeric RNA; cis-SAGe; genome-wide analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Differential regulatory roles of microRNAs during intramuscular adipogenesis in Chinese Guizhou Congjiang Xiang pigs.Epigenetics. 2022 Dec;17(12):1800-1819. doi: 10.1080/15592294.2022.2086675. Epub 2022 Jun 12. Epigenetics. 2022. PMID: 35695092 Free PMC article.
-
Novel read-through fusion transcript Bcl2l2-Pabpn1 in glioblastoma cells.J Cell Mol Med. 2022 Sep;26(17):4686-4697. doi: 10.1111/jcmm.17481. Epub 2022 Jul 27. J Cell Mol Med. 2022. PMID: 35894779 Free PMC article.
-
Construction of miRNA-mRNA network in the differentiation of chicken preadipocytes.Br Poult Sci. 2022 Jun;63(3):298-306. doi: 10.1080/00071668.2021.2000585. Epub 2022 Jan 14. Br Poult Sci. 2022. PMID: 34738495
-
Generation of Chimeric RNAs by cis-splicing of adjacent genes (cis-SAGe) in mammals.Yi Chuan. 2018 Feb 20;40(2):145-154. doi: 10.16288/j.yczz.17-197. Yi Chuan. 2018. PMID: 29428907 Review.
-
Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity.Oncotarget. 2016 Jun 28;7(26):40830-40845. doi: 10.18632/oncotarget.8518. Oncotarget. 2016. PMID: 27049726 Free PMC article. Review.
Cited by
-
Genome-wide association analysis of milk production, somatic cell score, and body conformation traits in Holstein cows.Front Vet Sci. 2022 Oct 4;9:932034. doi: 10.3389/fvets.2022.932034. eCollection 2022. Front Vet Sci. 2022. PMID: 36268046 Free PMC article.
-
Effects of HOXC8 on the Proliferation and Differentiation of Porcine Preadipocytes.Animals (Basel). 2023 Aug 14;13(16):2615. doi: 10.3390/ani13162615. Animals (Basel). 2023. PMID: 37627406 Free PMC article.
-
Genome-wide characterization of lncRNAs and mRNAs in muscles with differential intramuscular fat contents.Front Vet Sci. 2022 Aug 8;9:982258. doi: 10.3389/fvets.2022.982258. eCollection 2022. Front Vet Sci. 2022. PMID: 36003408 Free PMC article.
-
Trends and Prospects in Pig Genomics and Genetics.Genes (Basel). 2024 Sep 30;15(10):1292. doi: 10.3390/genes15101292. Genes (Basel). 2024. PMID: 39457416 Free PMC article.
-
Cancer fusion transcripts with human non-coding RNAs.Front Oncol. 2024 Jun 11;14:1415801. doi: 10.3389/fonc.2024.1415801. eCollection 2024. Front Oncol. 2024. PMID: 38919532 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

