Global MicroRNA Expression Profiling of Buffalo (Bubalus bubalis) Embryos at Different Developmental Stages Produced by Somatic Cell Nuclear Transfer and In-Vitro Fertilization Using RNA Sequencing
- PMID: 35328007
- PMCID: PMC8952793
- DOI: 10.3390/genes13030453
Global MicroRNA Expression Profiling of Buffalo (Bubalus bubalis) Embryos at Different Developmental Stages Produced by Somatic Cell Nuclear Transfer and In-Vitro Fertilization Using RNA Sequencing
Abstract
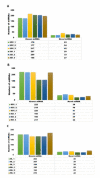
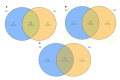
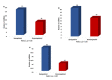
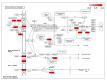
Despite the success of cloning technology in the production of offspring across several species, its application on a wide scale is severely limited by the very low offspring rate obtained with cloned embryos. The expression profile of microRNAs (miRNAs) in cloned embryos throughout embryonic development is reported to deviate from regular patterns. The present study is aimed at determining the dynamics of the global expression of miRNA profile in cloned and in-vitro fertilization (IVF) pre-implantation embryos at different developmental stages, i.e., the two-cell, eight-cell, and blastocyst stages, using next-generation sequencing. The results of this study suggest that there is a profound difference in global miRNA profile between cloned and IVF embryos. These differences are manifested throughout the course of embryonic development. The cloned embryos differ from their IVF counterparts in enriched Gene Ontology (GO) terms of biological process, molecular function, cellular component, and protein class categories in terms of the targets of differentially expressed miRNAs. The major pathways related to embryonic development, such as the Wnt signaling pathway, the apoptosis signaling pathway, the FGF signaling pathway, the p53 pathway, etc., were found to be affected in cloned relative to IVF embryos. Overall, these data reveal the distinct miRNA profile of cloned relative to IVF embryos, suggesting that the molecules or pathways affected may play an important role in cloned embryo development.
Keywords: In-vitro fertilization (IVF); RNA sequencing; bovine; gene ontology; miRNAs; somatic cell nuclear transfer (SCNT).
Conflict of interest statement
The authors do not have any conflict of interest.
Figures





Similar articles
-
Comparative transcriptome profile of embryos at different developmental stages derived from somatic cell nuclear transfer (SCNT) and in-vitro fertilization (IVF) in riverine buffalo (Bubalus bubalis).Vet Res Commun. 2024 Aug;48(4):2457-2475. doi: 10.1007/s11259-024-10419-8. Epub 2024 Jun 3. Vet Res Commun. 2024. PMID: 38829518
-
Selection of Reference miRNAs for Relative Quantification in Buffalo (Bubalus bubalis) Blastocysts Produced by Hand-Made Cloning and In Vitro Fertilization.Cell Reprogram. 2019 Aug;21(4):200-209. doi: 10.1089/cell.2019.0022. Epub 2019 Jun 14. Cell Reprogram. 2019. PMID: 31199674
-
RNA sequencing and transcriptome analysis of buffalo (Bubalus bubalis) blastocysts produced by somatic cell nuclear transfer and in vitro fertilization.Mol Reprod Dev. 2019 Sep;86(9):1149-1167. doi: 10.1002/mrd.23233. Epub 2019 Jul 15. Mol Reprod Dev. 2019. PMID: 31304661
-
Global DNA methylation profiles of buffalo (Bubalus bubalis) preimplantation embryos produced by handmade cloning and in vitro fertilization.Sci Rep. 2022 Mar 25;12(1):5161. doi: 10.1038/s41598-022-09207-8. Sci Rep. 2022. PMID: 35338228 Free PMC article.
-
An update: Reproductive handmade cloning of water buffalo (Bubalus bubalis).Anim Reprod Sci. 2018 Oct;197:1-9. doi: 10.1016/j.anireprosci.2018.08.003. Epub 2018 Aug 9. Anim Reprod Sci. 2018. PMID: 30122268 Review.
Cited by
-
Comparative transcriptome profile of embryos at different developmental stages derived from somatic cell nuclear transfer (SCNT) and in-vitro fertilization (IVF) in riverine buffalo (Bubalus bubalis).Vet Res Commun. 2024 Aug;48(4):2457-2475. doi: 10.1007/s11259-024-10419-8. Epub 2024 Jun 3. Vet Res Commun. 2024. PMID: 38829518
-
Effects of IGF-1 on the Three-Dimensional Culture of Ovarian Preantral Follicles and Superovulation Rates in Mice.Biology (Basel). 2022 May 29;11(6):833. doi: 10.3390/biology11060833. Biology (Basel). 2022. PMID: 35741354 Free PMC article.
-
RNA sequencing and gene co-expression network of in vitro matured oocytes and blastocysts of buffalo.Anim Reprod. 2024 Jun 17;21(2):e20230131. doi: 10.1590/1984-3143-AR2023-0131. eCollection 2024. Anim Reprod. 2024. PMID: 38912163 Free PMC article.
-
Molecular and cellular programs underlying the development of bovine pre-implantation embryos.Reprod Fertil Dev. 2023 Dec;36(2):34-42. doi: 10.1071/RD23146. Reprod Fertil Dev. 2023. PMID: 38064195 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

