Effect of the PmARF6 Gene from Masson Pine (Pinus massoniana) on the Development of Arabidopsis
- PMID: 35328022
- PMCID: PMC8949783
- DOI: 10.3390/genes13030469
Effect of the PmARF6 Gene from Masson Pine (Pinus massoniana) on the Development of Arabidopsis
Abstract
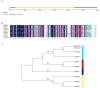
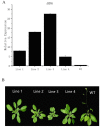
Masson pine (Pinus massoniana) is a core industrial tree species that is used for afforestation in southern China. Previous studies have shown that Auxin Response Factors (ARFs) are involved in the growth and development of various species, but the function of ARFs in Masson pine is unclear. In this research, we cloned and identified Masson pine ARF6 cDNA (PmARF6). The results showed that PmARF6 encodes a protein of 681 amino acids that is highly expressed in female flowers. Subcellular analysis showed that the PmARF6 protein occurred predominantly in the nucleus and cytomembrane of Masson pine cells. Compared with wild-type (WT) Arabidopsis, transgenic Arabidopsis plants overexpressing PmARF6 had fewer rosette leaves, and their flower development was slower. These results suggest that overexpression of PmARF6 may inhibit the flower and leaf development of Masson pine and provide new insights into the underlying developmental mechanism.
Keywords: Auxin Response Factor (ARF); Pinus massoniana; PmARF6; transgenic plant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yang Z., Xia H., Tan J., Feng Y., Huang Y. Selection of superior families of Pinus massoniana in southern China for large-diameter construction timber. J. For. Res. 2018;31:475–484. doi: 10.1007/s11676-018-0815-2. - DOI
-
- Bai Y., Zha X., Chen S. Effects of the vegetation restoration years on soil microbial community composition and biomass in degraded lands in Changting County, China. J. For. Res. 2019;31:1295–1308. doi: 10.1007/s11676-019-00879-z. - DOI
-
- Li C., Cheng P., Li Z., Xu Y., Sun Y., Qin D., Yu G. Transcriptomic and Metabolomic Analyses Provide Insights into the Enhancement of Torulene and Torularhodin Production in Rhodotorula glutinis ZHK under Moderate Salt Conditions. J. Agric. Food Chem. 2021;69:11523–11533. doi: 10.1021/acs.jafc.1c04028. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

