Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides
- PMID: 35328043
- PMCID: PMC8954616
- DOI: 10.3390/genes13030490
Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides
Abstract
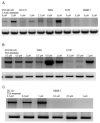
The ionotropic glutamate receptor 6 (GluR6 or GRIK2) gene is transcribed by two cell-type-specific promoters in neuronal and non-neuronal cells, which results in five different transcript variants. The purpose of this study was to explore cell-type-specific silencing of these promoters by epigenetic mechanisms. The neuronal and non-neuronal promoter sequences were cloned upstream of the luciferase gene in the pGL3 luciferase reporter vector. Promoter susceptibility to methylation was confirmed by 5-azacytidine and trichostatin treatment, and the status of CpG dinucleotides was determined by bisulfite sequencing of the promoter was determined by bisulfite sequences. GluR6A transcript variant was expressed in the brain, and GluR6B was most abundant in tumor cell lines. The neuronal promoter was methylated in non-neuronal cell lines. The treatment with 5-azacytidine and trichostatin upregulated transcription of the GluR6 gene, and methylation of the GluR6 promoter sequence in the luciferase reporter system led to downregulation of the luciferase gene transcription. Bisulfite sequencing revealed methylation of 3 and 41 CpG sites in non-neuronal and neuronal promoters, respectively. The differential activation/silencing of GluR6 promoters suggests that the transcript variants of GluR6 are involved in tissue-specific biological processes and their aberrant regulation in tumor cells may contribute to distinct properties of tumor cells.
Keywords: CpG methylation in GluR6 promoters; GRIK2; GluR6; GluR6 variants; bisulfite sequencing; carcinoma; epigenetic regulation; luciferase reporter; neuronal and non-neuronal promoters in GluR6/GRIK2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Lerma J. Ionotropic glutamate receptors in the CNS. In: Jonas P., Monyer H., editors. Handbook of Experimental Pharmacology. Volume 141. Springer; Berlin, Germany: 1999. pp. 275–307.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

