Complex of HIV-1 Integrase with Cellular Ku Protein: Interaction Interface and Search for Inhibitors
- PMID: 35328329
- PMCID: PMC8951179
- DOI: 10.3390/ijms23062908
Complex of HIV-1 Integrase with Cellular Ku Protein: Interaction Interface and Search for Inhibitors
Abstract
The interaction of HIV-1 integrase and the cellular Ku70 protein is necessary for HIV replication due to its positive effect on post-integration DNA repair. We have previously described in detail the Ku70 binding site within integrase. However, the integrase binding site in Ku70 remained poorly characterized. Here, using a peptide fishing assay and site-directed mutagenesis, we have identified residues I72, S73, and I76 of Ku70 as key for integrase binding. The molecular dynamics studies have revealed a possible way for IN to bind to Ku70, which is consistent with experimental data. According to this model, residues I72 and I76 of Ku70 form a "leucine zipper" with integrase residues, and, therefore, their concealment by low-molecular-weight compounds should impede the Ku70 interaction with integrase. We have identified such compounds by molecular docking and have confirmed their capacity to inhibit the formation of the integrase complex with Ku70. Our data demonstrate that the site of IN binding within Ku70 identified in the present work may be used for further search for inhibitors of the integrase binding to Ku70.
Keywords: HIV-1; Ku70; integrase; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


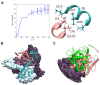

Similar articles
-
Characterization of HIV-1 integrase interaction with human Ku70 protein and initial implications for drug targeting.Sci Rep. 2017 Jul 17;7(1):5649. doi: 10.1038/s41598-017-05659-5. Sci Rep. 2017. PMID: 28717247 Free PMC article.
-
A Fluorescent Assay to Search for Inhibitors of HIV-1 Integrase Interactions with Human Ku70 Protein, and Its Application for Characterization of Oligonucleotide Inhibitors.Biomolecules. 2020 Aug 25;10(9):1236. doi: 10.3390/biom10091236. Biomolecules. 2020. PMID: 32854330 Free PMC article.
-
NHEJ pathway is involved in post-integrational DNA repair due to Ku70 binding to HIV-1 integrase.Retrovirology. 2019 Nov 6;16(1):30. doi: 10.1186/s12977-019-0492-z. Retrovirology. 2019. PMID: 31690330 Free PMC article.
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
In search of second-generation HIV integrase inhibitors: targeting integration beyond strand transfer.Future Med Chem. 2009 Oct;1(7):1259-74. doi: 10.4155/fmc.09.86. Future Med Chem. 2009. PMID: 21426102 Review.
Cited by
-
Involvement of Human Cellular Proteins and Structures in Realization of the HIV Life Cycle: A Comprehensive Review, 2024.Viruses. 2024 Oct 29;16(11):1682. doi: 10.3390/v16111682. Viruses. 2024. PMID: 39599797 Free PMC article. Review.
-
KuINins as a New Class of HIV-1 Inhibitors That Block Post-Integration DNA Repair.Int J Mol Sci. 2023 Dec 11;24(24):17354. doi: 10.3390/ijms242417354. Int J Mol Sci. 2023. PMID: 38139188 Free PMC article.
-
Association of Polymorphisms in NHEJ Pathway Genes with HIV-1 Infection and AIDS Progression in a Northern Chinese MSM Population.Dis Markers. 2022 Oct 19;2022:5126867. doi: 10.1155/2022/5126867. eCollection 2022. Dis Markers. 2022. PMID: 36312587 Free PMC article.
-
Recent Advances in Protein-Protein Interactions.Int J Mol Sci. 2023 Jan 9;24(2):1282. doi: 10.3390/ijms24021282. Int J Mol Sci. 2023. PMID: 36674795 Free PMC article.
-
Complex Relationships between HIV-1 Integrase and Its Cellular Partners.Int J Mol Sci. 2022 Oct 15;23(20):12341. doi: 10.3390/ijms232012341. Int J Mol Sci. 2022. PMID: 36293197 Free PMC article. Review.
References
-
- Günthard H.F., Calvez V., Paredes R., Pillay D., Shafer R.W., Wensing A.M., Jacobsen D.M., Richman D.D., Francisco S. Human Immunodeficiency Virus Drug Resistance: 2018 Recommendations of the International Antiviral Society-USA Panel. Clin. Infect. Dis. 2019;68:177–187. doi: 10.1093/cid/ciy463. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

