Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering
- PMID: 35328351
- PMCID: PMC8951411
- DOI: 10.3390/ijms23062928
Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering
Abstract
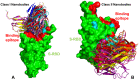
Nanobodies provide important advantages over traditional antibodies, including their smaller size and robust biochemical properties such as high thermal stability, high solubility, and the ability to be bioengineered into novel multivalent, multi-specific, and high-affinity molecules, making them a class of emerging powerful therapies against SARS-CoV-2. Recent research efforts on the design, protein engineering, and structure-functional characterization of nanobodies and their binding with SARS-CoV-2 S proteins reflected a growing realization that nanobody combinations can exploit distinct binding epitopes and leverage the intrinsic plasticity of the conformational landscape for the SARS-CoV-2 S protein to produce efficient neutralizing and mutation resistant characteristics. Structural and computational studies have also been instrumental in quantifying the structure, dynamics, and energetics of the SARS-CoV-2 spike protein binding with nanobodies. In this review, a comprehensive analysis of the current structural, biophysical, and computational biology investigations of SARS-CoV-2 S proteins and their complexes with distinct classes of nanobodies targeting different binding sites is presented. The analysis of computational studies is supplemented by an in-depth examination of mutational scanning simulations and identification of binding energy hotspots for distinct nanobody classes. The review is focused on the analysis of mechanisms underlying synergistic binding of multivalent nanobodies that can be superior to single nanobodies and conventional nanobody cocktails in combating escape mutations by effectively leveraging binding avidity and allosteric cooperativity. We discuss how structural insights and protein engineering approaches together with computational biology tools can aid in the rational design of synergistic combinations that exhibit superior binding and neutralization characteristics owing to avidity-mediated mechanisms.
Keywords: ACE2 host receptor; allosteric interactions; binding energy hotspots; biophysical methods; molecular dynamics; mutational scanning; signal transmission.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Allosteric Determinants of the SARS-CoV-2 Spike Protein Binding with Nanobodies: Examining Mechanisms of Mutational Escape and Sensitivity of the Omicron Variant.Int J Mol Sci. 2022 Feb 16;23(4):2172. doi: 10.3390/ijms23042172. Int J Mol Sci. 2022. PMID: 35216287 Free PMC article.
-
Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.Science. 2021 Feb 12;371(6530):eabe6230. doi: 10.1126/science.abe6230. Epub 2021 Jan 12. Science. 2021. PMID: 33436526 Free PMC article.
-
Nanobody engineering for SARS-CoV-2 neutralization and detection.Microbiol Spectr. 2024 Apr 2;12(4):e0419922. doi: 10.1128/spectrum.04199-22. Epub 2024 Feb 16. Microbiol Spectr. 2024. PMID: 38363137 Free PMC article.
-
Structural Biology of Nanobodies against the Spike Protein of SARS-CoV-2.Viruses. 2021 Nov 3;13(11):2214. doi: 10.3390/v13112214. Viruses. 2021. PMID: 34835020 Free PMC article. Review.
-
Nanobody engineering: computational modelling and design for biomedical and therapeutic applications.FEBS Open Bio. 2025 Feb;15(2):236-253. doi: 10.1002/2211-5463.13850. Epub 2024 Jun 19. FEBS Open Bio. 2025. PMID: 38898362 Free PMC article. Review.
Cited by
-
Nanobodies: From Discovery to AI-Driven Design.Biology (Basel). 2025 May 14;14(5):547. doi: 10.3390/biology14050547. Biology (Basel). 2025. PMID: 40427736 Free PMC article. Review.
-
Systematic surveillance of SARS-CoV-2 reveals dynamics of variant mutagenesis and transmission in a large urban population.Nat Commun. 2024 Dec 30;15(1):10795. doi: 10.1038/s41467-024-55031-1. Nat Commun. 2024. PMID: 39738001 Free PMC article.
-
Nanobodies: Robust miniprotein binders in biomedicine.Adv Drug Deliv Rev. 2023 Apr;195:114726. doi: 10.1016/j.addr.2023.114726. Epub 2023 Feb 7. Adv Drug Deliv Rev. 2023. PMID: 36754285 Free PMC article. Review.
-
Novel bispecific nanobody mitigates experimental intestinal inflammation in mice by targeting TNF-α and IL-23p19 bioactivities.Clin Transl Med. 2024 Mar;14(3):e1636. doi: 10.1002/ctm2.1636. Clin Transl Med. 2024. PMID: 38533646 Free PMC article.
-
Human ACE2 orthologous peptide sequences show better binding affinity to SARS-CoV-2 RBD domain: Implications for drug design.Comput Struct Biotechnol J. 2023 Jul 24;21:4096-4109. doi: 10.1016/j.csbj.2023.07.022. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37671240 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

