Tissue-Specific Methylation Biosignatures for Monitoring Diseases: An In Silico Approach
- PMID: 35328380
- PMCID: PMC8952417
- DOI: 10.3390/ijms23062959
Tissue-Specific Methylation Biosignatures for Monitoring Diseases: An In Silico Approach
Abstract
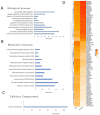
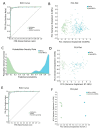
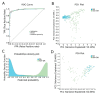
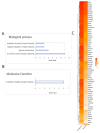
Tissue-specific gene methylation events are key to the pathogenesis of several diseases and can be utilized for diagnosis and monitoring. Here, we established an in silico pipeline to analyze high-throughput methylome datasets to identify specific methylation fingerprints in three pathological entities of major burden, i.e., breast cancer (BrCa), osteoarthritis (OA) and diabetes mellitus (DM). Differential methylation analysis was conducted to compare tissues/cells related to the pathology and different types of healthy tissues, revealing Differentially Methylated Genes (DMGs). Highly performing and low feature number biosignatures were built with automated machine learning, including: (1) a five-gene biosignature discriminating BrCa tissue from healthy tissues (AUC 0.987 and precision 0.987), (2) three equivalent OA cartilage-specific biosignatures containing four genes each (AUC 0.978 and precision 0.986) and (3) a four-gene pancreatic β-cell-specific biosignature (AUC 0.984 and precision 0.995). Next, the BrCa biosignature was validated using an independent ccfDNA dataset showing an AUC and precision of 1.000, verifying the biosignature's applicability in liquid biopsy. Functional and protein interaction prediction analysis revealed that most DMGs identified are involved in pathways known to be related to the studied diseases or pointed to new ones. Overall, our data-driven approach contributes to the maximum exploitation of high-throughput methylome readings, helping to establish specific disease profiles to be applied in clinical practice and to understand human pathology.
Keywords: breast cancer; diabetes; liquid biopsy; machine learning; methylation; microarrays; model; osteoarthritis.
Conflict of interest statement
I.T. is the CEO and founder of JADBio.
Figures



Similar articles
-
Deciphering the Methylation Landscape in Breast Cancer: Diagnostic and Prognostic Biosignatures through Automated Machine Learning.Cancers (Basel). 2021 Apr 2;13(7):1677. doi: 10.3390/cancers13071677. Cancers (Basel). 2021. PMID: 33918195 Free PMC article.
-
Mitochondrial Fraction of Circulating Cell-Free DNA as an Indicator of Human Pathology.Int J Mol Sci. 2024 Apr 10;25(8):4199. doi: 10.3390/ijms25084199. Int J Mol Sci. 2024. PMID: 38673785 Free PMC article.
-
Liquid Biopsy in Type 2 Diabetes Mellitus Management: Building Specific Biosignatures via Machine Learning.J Clin Med. 2022 Feb 17;11(4):1045. doi: 10.3390/jcm11041045. J Clin Med. 2022. PMID: 35207316 Free PMC article.
-
DNA methylation biomarkers for noninvasive detection of triple-negative breast cancer using liquid biopsy.Int J Cancer. 2023 Mar 1;152(5):1025-1035. doi: 10.1002/ijc.34337. Epub 2022 Nov 8. Int J Cancer. 2023. PMID: 36305646 Review.
-
The Epigenomic Landscape in Osteoarthritis.Curr Rheumatol Rep. 2017 Jun;19(6):30. doi: 10.1007/s11926-017-0661-9. Curr Rheumatol Rep. 2017. PMID: 28456906 Free PMC article. Review.
Cited by
-
Prediction and Ranking of Biomarkers Using multiple UniReD.Int J Mol Sci. 2022 Sep 21;23(19):11112. doi: 10.3390/ijms231911112. Int J Mol Sci. 2022. PMID: 36232413 Free PMC article.
-
A novel blood-based epigenetic biosignature in first-episode schizophrenia patients through automated machine learning.Transl Psychiatry. 2024 Jun 17;14(1):257. doi: 10.1038/s41398-024-02946-4. Transl Psychiatry. 2024. PMID: 38886359 Free PMC article.
-
Automated machine learning for genome wide association studies.Bioinformatics. 2023 Sep 2;39(9):btad545. doi: 10.1093/bioinformatics/btad545. Bioinformatics. 2023. PMID: 37672022 Free PMC article.
-
Label-Free Human Disease Characterization through Circulating Cell-Free DNA Analysis Using Raman Spectroscopy.Int J Mol Sci. 2023 Aug 3;24(15):12384. doi: 10.3390/ijms241512384. Int J Mol Sci. 2023. PMID: 37569759 Free PMC article.
-
Tracing the Origin of Cell-Free DNA Molecules through Tissue-Specific Epigenetic Signatures.Diagnostics (Basel). 2022 Jul 29;12(8):1834. doi: 10.3390/diagnostics12081834. Diagnostics (Basel). 2022. PMID: 36010184 Free PMC article. Review.
References
-
- Kulis M., Esteller M. 2–DNA Methylation and Cancer. In: Herceg Z., Ushijima T., editors. Advances in Genetics. Volume 70. Academic Press; Cambridge, MA, USA: 2010. pp. 27–56. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

