DNMTs Are Involved in TGF-β1-Induced Epithelial-Mesenchymal Transitions in Airway Epithelial Cells
- PMID: 35328422
- PMCID: PMC8951572
- DOI: 10.3390/ijms23063003
DNMTs Are Involved in TGF-β1-Induced Epithelial-Mesenchymal Transitions in Airway Epithelial Cells
Abstract
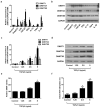
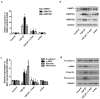
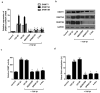
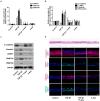
Chronic rhinosinusitis (CRS) pathogenesis is closely related to tissue remodeling, including epithelial-mesenchymal transition (EMT). Epigenetic mechanisms play key roles in EMT. DNA methylation, mediated by DNA methyltransferases (DNMTs), is an epigenetic marker that is critical to EMT. The goal of this study was to determine whether DNMTs were involved in TGF-β1-induced EMT and elucidate the underlying mechanisms in nasal epithelial cells and air-liquid interface cultures. Global DNA methylation and DNMT activity were quantified. DNMT expression was measured using real-time PCR (qRT-PCR) in human CRS tissues. mRNA and protein levels of DNMTs, E-cadherin, vimentin, α-SMA, and fibronectin were determined using RT-PCR and Western blotting, respectively. DNMT1, DNMT3A, and DNMT3B gene expression were knocked down using siRNA transfection. MAPK phosphorylation and EMT-related transcription factor levels were determined using Western blotting. Signaling pathways were analyzed using specific inhibitors of MAPK. We demonstrated these data in primary nasal epithelial cells and air-liquid interface cultures. Global DNA methylation, DNMT activity, and DNMT expression increased in CRS tissues. DNMT expression was positively correlated with Lund-McKay CT scores. TGF-β1 dose-dependently induced DNMT expression. Further, 5-Aza inhibited TGF-β1-induced DNMT, Snail, and Slug expression related to EMT, as well as p38 and JNK phosphorylation in A549 cells and TGF-β1-induced DNMT expression and EMT in primary nasal epithelial cells and air-liquid interface cultures. TGF-β1-induced DNMT expression leads to DNA methylation and EMT via p38, JNK, Snail, and Slug signaling pathways. Inhibition of DNMT suppressed the EMT process and therefore is potentially a CRS therapeutic strategy.
Keywords: DNA methyltransferase; epithelial–mesenchymal transition; primary nasal epithelial cells; tissue remodeling; transforming growth factor β-1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Trichostatin A Inhibits Epithelial Mesenchymal Transition Induced by TGF-β1 in Airway Epithelium.PLoS One. 2016 Aug 29;11(8):e0162058. doi: 10.1371/journal.pone.0162058. eCollection 2016. PLoS One. 2016. PMID: 27571418 Free PMC article.
-
miR-29b Regulates TGF-β1-Induced Epithelial-Mesenchymal Transition by Inhibiting Heat Shock Protein 47 Expression in Airway Epithelial Cells.Int J Mol Sci. 2021 Oct 26;22(21):11535. doi: 10.3390/ijms222111535. Int J Mol Sci. 2021. PMID: 34768968 Free PMC article.
-
TGF-β1 Induces Epithelial-Mesenchymal Transition of Chronic Sinusitis with Nasal Polyps through MicroRNA-21.Int Arch Allergy Immunol. 2019;179(4):304-319. doi: 10.1159/000497829. Epub 2019 Apr 12. Int Arch Allergy Immunol. 2019. PMID: 30982052
-
The Role of Epithelial-Mesenchymal Transition in Chronic Rhinosinusitis.Int Arch Allergy Immunol. 2022;183(10):1029-1039. doi: 10.1159/000524950. Epub 2022 Jun 23. Int Arch Allergy Immunol. 2022. PMID: 35738243 Review.
-
Epithelial cell dysfunction in chronic rhinosinusitis: the epithelial-mesenchymal transition.Expert Rev Clin Immunol. 2023 Jul-Dec;19(8):959-968. doi: 10.1080/1744666X.2023.2232113. Epub 2023 Jul 4. Expert Rev Clin Immunol. 2023. PMID: 37386882 Review.
Cited by
-
Alveolar epithelial cell dysfunction and epithelial-mesenchymal transition in pulmonary fibrosis pathogenesis.Front Mol Biosci. 2025 Apr 24;12:1564176. doi: 10.3389/fmolb.2025.1564176. eCollection 2025. Front Mol Biosci. 2025. PMID: 40343260 Free PMC article. Review.
-
Evaluation of CNPase and TGFβ1/Smad Signalling Pathway Molecule Expression in Sinus Epithelial Tissues of Patients with Chronic Rhinosinusitis with (CRSwNP) and without Nasal Polyps (CRSsNP).J Pers Med. 2024 Aug 23;14(9):894. doi: 10.3390/jpm14090894. J Pers Med. 2024. PMID: 39338148 Free PMC article.
-
5-Aza-2'-Deoxycytidine (5-Aza-dC, Decitabine) Inhibits Collagen Type I and III Expression in TGF-β1-Treated Equine Endometrial Fibroblasts.Animals (Basel). 2023 Mar 30;13(7):1212. doi: 10.3390/ani13071212. Animals (Basel). 2023. PMID: 37048467 Free PMC article.
-
Epithelial-Mesenchymal Transition in Chronic Rhinosinusitis.J Rhinol. 2024 Jul;31(2):67-77. doi: 10.18787/jr.2024.00022. Epub 2024 Jul 31. J Rhinol. 2024. PMID: 39664411 Free PMC article. Review.
-
Drug tolerance and persistence to EGFR inhibitor treatment are mediated by an ILK-SFK-YAP signaling axis in lung adenocarcinoma.Oncogene. 2025 Aug;44(32):2831-2849. doi: 10.1038/s41388-025-03461-6. Epub 2025 May 31. Oncogene. 2025. PMID: 40450112 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

