Identifying Function Determining Residues in Neuroimmune Semaphorin 4A
- PMID: 35328445
- PMCID: PMC8953949
- DOI: 10.3390/ijms23063024
Identifying Function Determining Residues in Neuroimmune Semaphorin 4A
Abstract
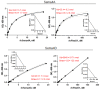
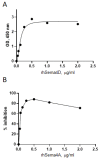
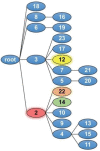
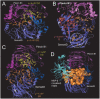
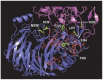
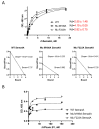
Semaphorin 4A (Sema4A) exerts a stabilizing effect on human Treg cells in PBMC and CD4+ T cell cultures by engaging Plexin B1. Sema4A deficient mice display enhanced allergic airway inflammation accompanied by fewer Treg cells, while Sema4D deficient mice displayed reduced inflammation and increased Treg cell numbers even though both Sema4 subfamily members engage Plexin B1. The main objectives of this study were: 1. To compare the in vitro effects of Sema4A and Sema4D proteins on human Treg cells; and 2. To identify function-determining residues in Sema4A critical for binding to Plexin B1 based on Sema4D homology modeling. We report here that Sema4A and Sema4D display opposite effects on human Treg cells in in vitro PBMC cultures; Sema4D inhibited the CD4+CD25+Foxp3+ cell numbers and CD25/Foxp3 expression. Sema4A and Sema4D competitively bind to Plexin B1 in vitro and hence may be doing so in vivo as well. Bayesian Partitioning with Pattern Selection (BPPS) partitioned 4505 Sema domains from diverse organisms into subgroups based on distinguishing sequence patterns that are likely responsible for functional differences. BPPS groups Sema3 and Sema4 into one family and further separates Sema4A and Sema4D into distinct subfamilies. Residues distinctive of the Sema3,4 family and of Sema4A (and by homology of Sema4D) tend to cluster around the Plexin B1 binding site. This suggests that the residues both common to and distinctive of Sema4A and Sema4D may mediate binding to Plexin B1, with subfamily residues mediating functional specificity. We mutated the Sema4A-specific residues M198 and F223 to alanine; notably, F223 in Sema4A corresponds to alanine in Sema4D. Mutant proteins were assayed for Plexin B1-binding and Treg stimulation activities. The F223A mutant was unable to stimulate Treg stability in in vitro PBMC cultures despite binding Plexin B1 with an affinity similar to the WT protein. This research is a first step in generating potent mutant Sema4A molecules with stimulatory function for Treg cells with a view to designing immunotherapeutics for asthma.
Keywords: Plexin B1; Semaphorin 4A; human Treg cells; immunotherapeutics for asthma; mutated proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Semaphorin 4A Stabilizes Human Regulatory T Cell Phenotype via Plexin B1.Immunohorizons. 2019 Feb;3(2):71-87. doi: 10.4049/immunohorizons.1800026. Immunohorizons. 2019. PMID: 31236543 Free PMC article.
-
Expression of neuroimmune semaphorins 4A and 4D and their receptors in the lung is enhanced by allergen and vascular endothelial growth factor.BMC Immunol. 2011 May 19;12:30. doi: 10.1186/1471-2172-12-30. BMC Immunol. 2011. PMID: 21595947 Free PMC article.
-
Class 4 Semaphorins and Plexin-B receptors regulate GABAergic and glutamatergic synapse development in the mammalian hippocampus.Mol Cell Neurosci. 2018 Oct;92:50-66. doi: 10.1016/j.mcn.2018.06.008. Epub 2018 Jul 4. Mol Cell Neurosci. 2018. PMID: 29981480 Free PMC article.
-
The Plexin-B family and its role in cancer progression.Histol Histopathol. 2014 Feb;29(2):151-65. doi: 10.14670/HH-29.151. Epub 2013 Sep 17. Histol Histopathol. 2014. PMID: 24043639 Review.
-
Biology and function of neuroimmune semaphorins 4A and 4D.Immunol Res. 2011 May;50(1):10-21. doi: 10.1007/s12026-010-8201-y. Immunol Res. 2011. PMID: 21203905 Free PMC article. Review.
Cited by
-
Plexin B1 controls Treg numbers, limits allergic airway inflammation, and regulates mucins.Front Immunol. 2024 Jan 8;14:1297354. doi: 10.3389/fimmu.2023.1297354. eCollection 2023. Front Immunol. 2024. PMID: 38259471 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

