Discovery and Biotechnological Exploitation of Glycoside-Phosphorylases
- PMID: 35328479
- PMCID: PMC8950772
- DOI: 10.3390/ijms23063043
Discovery and Biotechnological Exploitation of Glycoside-Phosphorylases
Abstract
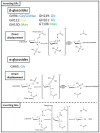
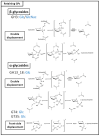
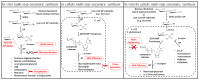
Among carbohydrate active enzymes, glycoside phosphorylases (GPs) are valuable catalysts for white biotechnologies, due to their exquisite capacity to efficiently re-modulate oligo- and poly-saccharides, without the need for costly activated sugars as substrates. The reversibility of the phosphorolysis reaction, indeed, makes them attractive tools for glycodiversification. However, discovery of new GP functions is hindered by the difficulty in identifying them in sequence databases, and, rather, relies on extensive and tedious biochemical characterization studies. Nevertheless, recent advances in automated tools have led to major improvements in GP mining, activity predictions, and functional screening. Implementation of GPs into innovative in vitro and in cellulo bioproduction strategies has also made substantial advances. Herein, we propose to discuss the latest developments in the strategies employed to efficiently discover GPs and make the best use of their exceptional catalytic properties for glycoside bioproduction.
Keywords: biotechnology; carbohydrate-active enzymes; functional metagenomics; glycochemistry; glycoside phosphorylases; glycosides; screening; sequence similarity networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rudroff F., Mihovilovic M.D., Gröger H., Snajdrova R., Iding H., Bornscheuer U.T. Opportunities and challenges for combining chemo- and biocatalysis. Nat. Catal. 2018;1:12–22. doi: 10.1038/s41929-017-0010-4. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

