Blood Transcriptome Profiling Links Immunity to Disease Severity in Myotonic Dystrophy Type 1 (DM1)
- PMID: 35328504
- PMCID: PMC8954763
- DOI: 10.3390/ijms23063081
Blood Transcriptome Profiling Links Immunity to Disease Severity in Myotonic Dystrophy Type 1 (DM1)
Abstract
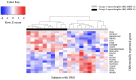
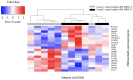
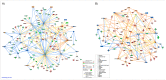
The blood transcriptome was examined in relation to disease severity in type I myotonic dystrophy (DM1) patients who participated in the Observational Prolonged Trial In DM1 to Improve QoL- Standards (OPTIMISTIC) study. This sought to (a) ascertain if transcriptome changes were associated with increasing disease severity, as measured by the muscle impairment rating scale (MIRS), and (b) establish if these changes in mRNA expression and associated biological pathways were also observed in the Dystrophia Myotonica Biomarker Discovery Initiative (DMBDI) microarray dataset in blood (with equivalent MIRS/DMPK repeat length). The changes in gene expression were compared using a number of complementary pathways, gene ontology and upstream regulator analyses, which suggested that symptom severity in DM1 was linked to transcriptomic alterations in innate and adaptive immunity associated with muscle-wasting. Future studies should explore the role of immunity in DM1 in more detail to assess its relevance to DM1.
Keywords: DM1 disease severity; RNA sequencing; blood; immunity; muscle impairment rating scale (MIRS); myotonic dystrophy type 1 (DM1); pathway analysis.
Conflict of interest statement
The authors declare that they have no conflict of interest except van Engelen. van Engelen reports grants from FP7 European Union grant OPTIMISTIC, grants from Marigold Canada, grants from Spieren voor Spieren, grants from the Prinses Beatrix Spierfonds, grants from the Dutch FSHD foundation, grants from the Netherlands Organisation for Scientific Research (NWO), grants from the Netherlands Organisation for Health Research and Development (ZonMw), grants from Association Francaise contre les Myopathies, and grants and personal fees from Fulcrum and Facio, outside the submitted work.
Figures





References
-
- Bird T.D. Myotonic Dystrophy Type 1. In: Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Ledbetter N., Mefford H.C., Smith R.J.H., et al., editors. GeneReviews(R) University of Washington; Seattle, WA, USA: 1993. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

